| (1) AGO2.ip | (3) B-CELL | (3) BRAIN | (8) BREAST | (20) CELL-LINE | (3) CERVIX | (2) HEART | (4) LIVER | (2) OTHER | (6) SKIN | (1) TESTES | (1) UTERUS | (1) XRN.ip |
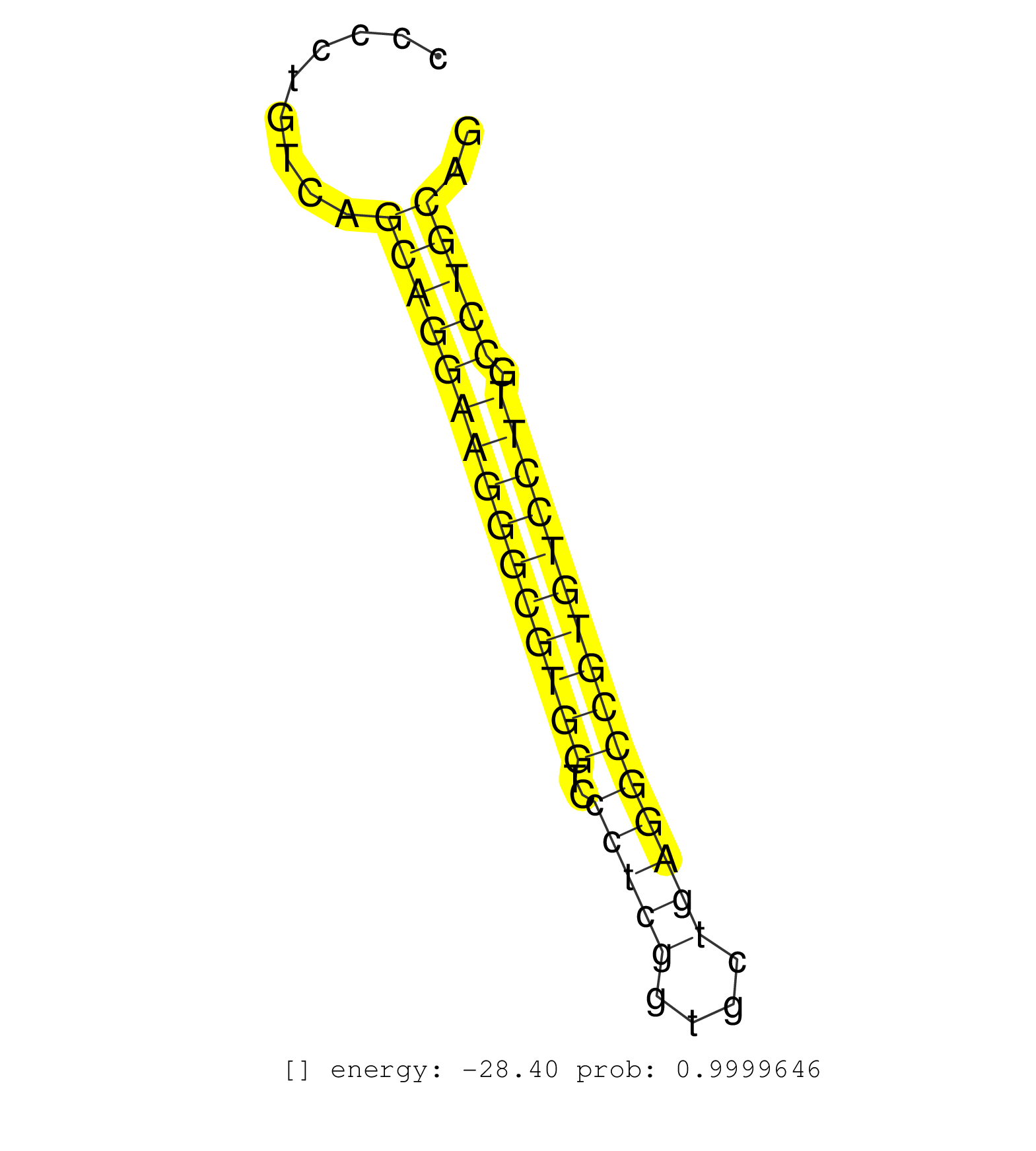
| GAAGAATTCTCCCAGCCTGCACCCACCTCAGAGAATTGGTTCATGAGGGGCCAGCCTGTCATGCAAAATCCAACTGCTGATTGACTAAGCAAGCATCAGCAGTGGCAGGCGCCATCCCATTGTGTGGAGCTCCCCACCCCCACCCCTGTCAGCAGGAAGGGCGTGGTCCCTCGGTGCTGAGGCCGTGTCCTTGCCTGCAGCCATCGAAGCCATGAAGAACGCCCACCGGGAGGAAATGGAGCGGGAGCTG .......................................................................................................................................................(((((((((((((((..(((((....))))))))))))))).))))).................................................... ..............................................................................................................................................143......................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553576(SRX182782) source: Testis. (testes) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189787 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR038857(GSM458540) D20. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR038859(GSM458542) MM386. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | GSM532878(GSM532878) G691T. (cervix) | SRR390723(GSM850202) total small RNA. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR029126(GSM416755) 143B. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR444063(SRX128911) Sample 20cDNABarcode: AF-PP-333: ACG CTC TTC . (cell line) | SRR189784 | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | TAX577745(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGGTC.................................................................................. | 21 | 1 | 19.00 | 19.00 | - | 8.00 | 6.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGTCC................................................................................. | 21 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CCCACCGGGAGGAAATGGAGC........ | 21 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGGTCA................................................................................. | 22 | 1 | 3.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGAAGAACGCCCACCGGGAGGAAATGGAGC........ | 30 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGG.................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGGAAT................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGT................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CAGCAGGAAGGGCGTGGTCCAT............................................................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................AGGCCGTGTCCTTGCCTGCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGTAT................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGG.................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGCCC................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CCCACCGGGAGGAAATGGAGCGGGAGC.. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GGAAGGGCGTGGTCCCTAA............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................TGAAGAACGCCCACCGGGAGGAAAT............. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGGTCT................................................................................. | 22 | 1 | 1.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGAGGCCGTGTCCTTGCCTCT.................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGTCCCCT.............................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGATA.................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................TCGAAGCCATGAAGAACGCCCACCG...................... | 25 | 2 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGGCCGTGTCCTTGCCTGCAGT................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGAAGAACGCCCACCGGGAGGAAATGG........... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGTCCA................................................................................ | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGGCCGTGTCCTTGCCTGCAGTAG............................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ACCGGGAGGAAATGGAGGCA...... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................GAAGCCATGAAGAACTACA.......................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................CAGCAGGAAGGGCGTGGAATA................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCAGCAGGAAGGGCGTGGAA.................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CAGCAGGAAGGGCGTGGTC.................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGAGGGGCCAGCCTGTCATGCAAAACCC................................................................................................................................................................................... | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTAGTC.................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................AACGCCCACCGGGAGGAAAT............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CAGCAGGAAGGGCGTGG.................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................AGGAAGGGCGTGGTCGGTG.............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................TGAAGAACGCCCACCGGGAGGAAATGGA.......... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GCAGGAAGGGCGTGGTC.................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTCAGCAGGAAGGGCGTGGTCTAT............................................................................... | 24 | 1 | 1.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGCCATCGAAGCCATGAAGAA............................... | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GAGGAAATGGAGCGGGACA.. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................CGGGAGGAAATGGAGC........ | 16 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 |
| ...............................................................................................................................................................................................................AGCCATGAAGAACGCCCACCGGGAG.................. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CCATGAAGAACGCCCACCGGGAGG................. | 24 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CGAAGCCATGAAGAACGCCCACCG...................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TCGAAGCCATGAAGAACGCCCACCGG..................... | 26 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................CATGAAGAACGCCCACCGGGAGG................. | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..................................................................................................................................................................................................................CATGAAGAACGCCCACCGG..................... | 19 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATGAAGAACGCCCACCGGG.................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CATCGAAGCCATGAAGAACGCCCAC........................ | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................................................................................................................AGCCATCGAAGCCATGAAG................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATGAAGAACGCCCACCGGGAG.................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATCGAAGCCATGAAGAACGCCCACCGGGAGG................. | 31 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGAAGAACGCCCACCGG..................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATGAAGAACGCCCACC....................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAAGAATTCTCCCAGCCTGCACCCACCTCAGAGAATTGGTTCATGAGGGGCCAGCCTGTCATGCAAAATCCAACTGCTGATTGACTAAGCAAGCATCAGCAGTGGCAGGCGCCATCCCATTGTGTGGAGCTCCCCACCCCCACCCCTGTCAGCAGGAAGGGCGTGGTCCCTCGGTGCTGAGGCCGTGTCCTTGCCTGCAGCCATCGAAGCCATGAAGAACGCCCACCGGGAGGAAATGGAGCGGGAGCTG .......................................................................................................................................................(((((((((((((((..(((((....))))))))))))))).))))).................................................... ..............................................................................................................................................143......................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553576(SRX182782) source: Testis. (testes) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189787 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR038857(GSM458540) D20. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR038859(GSM458542) MM386. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | GSM532878(GSM532878) G691T. (cervix) | SRR390723(GSM850202) total small RNA. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR029126(GSM416755) 143B. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR444063(SRX128911) Sample 20cDNABarcode: AF-PP-333: ACG CTC TTC . (cell line) | SRR189784 | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | TAX577745(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..AGAATTCTCCCAGCCTCTG..................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........TCCCAGCCTGCACCCACCTCAG........................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GAGCTCCCCACCCCCACTG......................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TGAGGCCGTGTCCTTTCCG...................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GCCATCGAAGCCATGAAGAACGCCCA......................... | 26 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CATCGAAGCCATGAAGA................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |