| (3) AGO2.ip | (6) B-CELL | (2) BRAIN | (8) BREAST | (22) CELL-LINE | (3) CERVIX | (5) HEART | (1) HELA | (4) LIVER | (1) OTHER | (16) SKIN | (1) TESTES | (3) UTERUS | (1) XRN.ip |
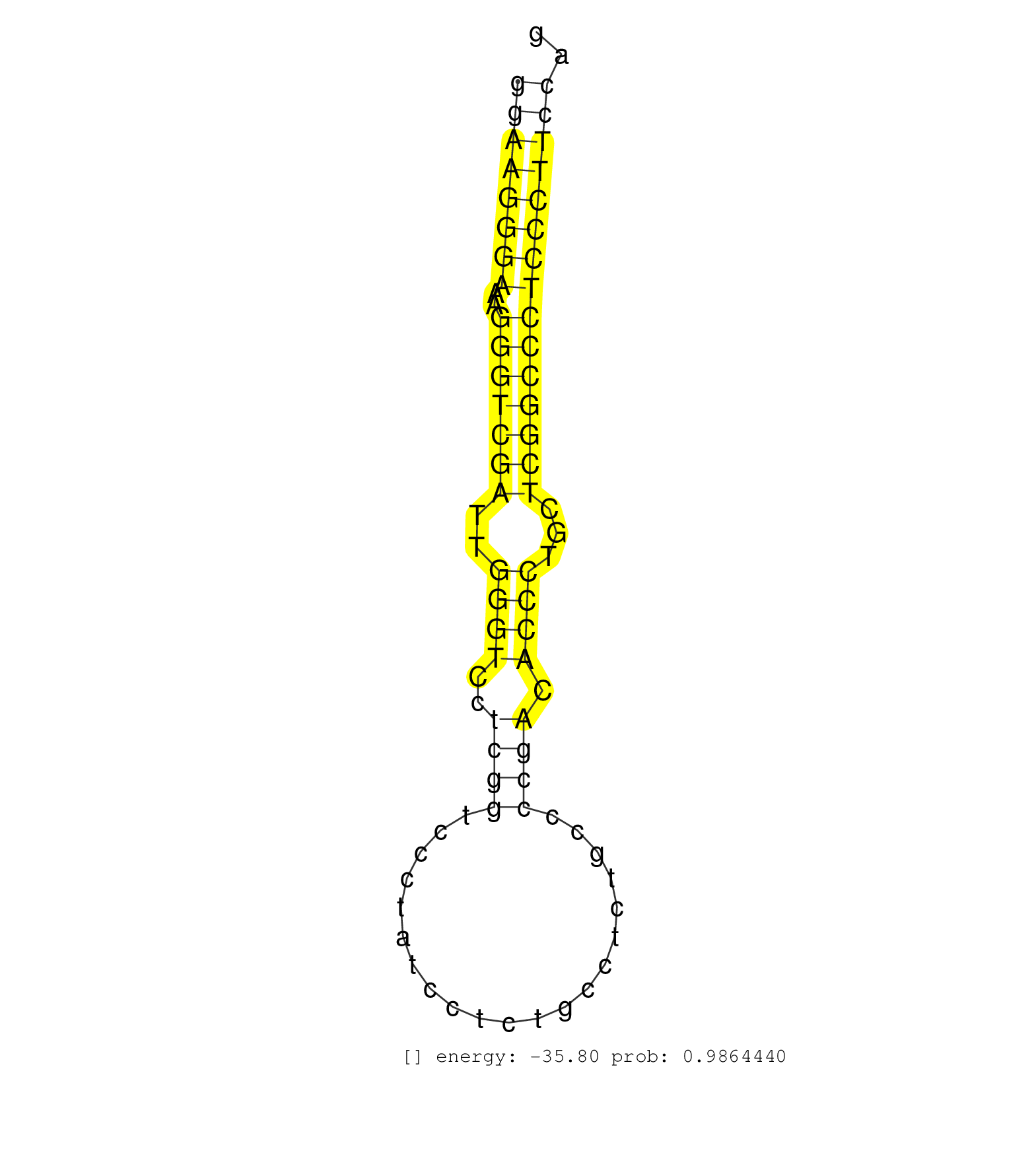
| CTGGTGAGGATGCAGATAGAGGCCAGCACTCCCAGCTTCCTGTGGGGCTAGGGCTCCTGCCTATTTCTTCTGGGCTGAGCTCTGCTAAACCTATGACCACCCTGTCAGAGCCAGAGGAGTAGGAAGGGAAAGGGTCGATTGGGTCCTCGGTCCCTATCCTCTGCCTCTGCCCCGACACCCTGCTCGGCCCTCCCTTCCAGACTTGGGCCTCTTCTCCACCAAGACCCTGGTGGAAGCGAGTGGCGAACAC .........................................................................................................................((((((((..(((((((..((((..((((.....................)))).))))...))))))))))))))).................................................... .........................................................................................................................122...........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR189782 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR189784 | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343336 | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR343334 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR040010(GSM532895) G529N. (cervix) | SRR444042(SRX128890) Sample 3cDNABarcode: AF-PP-335: ACG CTC TTC C. (skin) | SRR189783 | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577589(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191504(GSM715614) 124genomic small RNA (size selected RNA from . (breast) | SRR343337 | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR189785 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR038861(GSM458544) MM466. (cell line) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | SRR029131(GSM416760) MCF7. (cell line) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR444043(SRX128891) Sample 4cDNABarcode: AF-PP-339: ACG CTC TTC C. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................TGTGGGGCTAGGGCTAACA............................................................................................................................................................................................... | 19 | 13.00 | 0.00 | - | 5.00 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................GGAAAGGGTCGATTGGGT.......................................................................................................... | 18 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGTC......................................................................................................... | 22 | 1 | 9.00 | 9.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGT.......................................................................................................... | 21 | 1 | 6.00 | 6.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGGT.......................................................................................................... | 24 | 1 | 6.00 | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGTGGGGCTAGGGCTAAC................................................................................................................................................................................................ | 18 | 5.00 | 0.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTT...................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AGGGAAAGGGTCGATTGGGTCA........................................................................................................ | 22 | 4.00 | 0.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTGCTCGGCCCTCCCTTCCAGT................................................. | 22 | 1 | 4.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................GCGAGTGGCGAAC.. | 13 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGGTCCCTATCCTCTGCGCGC.................................................................................. | 22 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTTC..................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGTCCTTT..................................................................................................... | 26 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGTAA........................................................................................................ | 23 | 1 | 3.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGGTCCCTATCCTCTGCGCG................................................................................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGTA......................................................................................................... | 22 | 1 | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGCTCGGCCCTCCCTTCCAGA................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGCTCGGCCCTCCCTTCCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGTGGGGCTAGGGCTCACAG.............................................................................................................................................................................................. | 20 | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................CGGTCCCTATCCTCTGCGC.................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGG........................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGA........................................................................................................... | 23 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTG............................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CGGTCCCTATCCTCTGT...................................................................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................GGTGGAAGCGAGTGGCGAAC.. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTTCT.................................................... | 24 | 1 | 2.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TTCCTGTGGGGCTAGGGC.................................................................................................................................................................................................... | 18 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCT......................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CCCTATCCTCTGCCTCTGCCCCGACACC....................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AGGGAAAGGGTCGATTGGGTCC........................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TGGTGGAAGCGAGTGGCGA.... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GGGCCTCTTCTCCACCAAGACC........................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AAGGGTCGATTGGGTCCTCGGT................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCTCGGCCCTCCCTTCCAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGCTCGGCCCTCCCTTCCA................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AAGGGTCGATTGGGTCCTT...................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................AGGGAAAGGGTCGATTGGGTCCTC...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGGTT......................................................................................................... | 25 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGGG.......................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................TGTGGGGCTAGGGCTAT................................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................TTGGGCCTCTTCTCCACCAAGACCC....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGA................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGG............................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAGGGAAAGGGTCGATTGGGTCC........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTC.......................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTTCA.................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AGACCCTGGTGGAAGCGAG.......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGCTCGGCCCTCCCTTCCAA.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AGGATGCAGATAGAGGC................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................CTGGTGGAAGCGAGTGGCGA.... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGGCCTCTTCTCCACCAAGACCC....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................GGTGGAAGCGAGTGGCGAACA. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGAAAT........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GTGGAAGCGAGTGGCGAACA. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAGGGAAAGGGTCGATT.............................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGAAGGGAAAGGGTCGATTGGGTTT........................................................................................................ | 26 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TCTCCACCAAGACCCTGGT................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CACCCTGCTCGGCCCTCCCTTCC.................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGTGGGGCTAGGGCTATAT............................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................AGACCCTGGTGGAAGCGAGTGGCG..... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGTGGGGCTAGGGCTAAA................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................TGTGGGGCTAGGGCTAACG............................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................TGGTGGAAGCGAGTGGCGC.... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTTCC.................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCAGAGCCAGAGGAGTAGGAAGG........................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AAAGGGTCGATTGGGTCCTCGGT................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CCTGGTGGAAGCGAGTGG....... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTTTAT....................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACACCCTGCTCGGCCCTCCCTTA..................................................... | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................CGAGTGGCGAAC.. | 12 | 5 | 0.60 | 0.60 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| .................................................................................................................................AAGGGTCGATTGGGT.......................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .................................................................................................CACCCTGTCAGAGCCA......................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ......................................CCTGTGGGGCTAGGGC.................................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GGGCCTCTTCTCCACC.............................. | 16 | 8 | 0.12 | 0.12 | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACACCCTGCTCGGC.............................................................. | 14 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - |
| .................................................................TCTTCTGGGCTGAGC.......................................................................................................................................................................... | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| CTGGTGAGGATGCAGATAGAGGCCAGCACTCCCAGCTTCCTGTGGGGCTAGGGCTCCTGCCTATTTCTTCTGGGCTGAGCTCTGCTAAACCTATGACCACCCTGTCAGAGCCAGAGGAGTAGGAAGGGAAAGGGTCGATTGGGTCCTCGGTCCCTATCCTCTGCCTCTGCCCCGACACCCTGCTCGGCCCTCCCTTCCAGACTTGGGCCTCTTCTCCACCAAGACCCTGGTGGAAGCGAGTGGCGAACAC .........................................................................................................................((((((((..(((((((..((((..((((.....................)))).))))...))))))))))))))).................................................... .........................................................................................................................122...........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR189782 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR189784 | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343336 | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR343334 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR040010(GSM532895) G529N. (cervix) | SRR444042(SRX128890) Sample 3cDNABarcode: AF-PP-335: ACG CTC TTC C. (skin) | SRR189783 | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577589(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191504(GSM715614) 124genomic small RNA (size selected RNA from . (breast) | SRR343337 | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR189785 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR038861(GSM458544) MM466. (cell line) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | SRR029131(GSM416760) MCF7. (cell line) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR444043(SRX128891) Sample 4cDNABarcode: AF-PP-339: ACG CTC TTC C. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................TCCCTTCCAGACTTGTAAA......................................... | 19 | 2.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................GGCCAGCACTCCCAGCTTCCTGTCCT............................................................................................................................................................................................................ | 26 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................GCCTATTTCTTCTGGG................................................................................................................................................................................ | 16 | 5 | 1.20 | 1.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | 0.20 | 0.20 | 0.20 | - | - |
| .........................................................................................................................................................................................................CTTGGGCCTCTTCTCCGAC.............................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................CCTCTGCCCCGACACTTTT.................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................ACTCCCAGCTTCCTGTCAGC........................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..GGTGAGGATGCAGATAGTTCT................................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TCCCTTCCAGACTTGGTA.......................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................GGGCCTCTTCTCCACCAATGG......................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................GCCTATTTCTTCTGG................................................................................................................................................................................. | 15 | 10 | 0.60 | 0.60 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CACCCTGCTCGGCCC............................................................ | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |