| (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (5) B-CELL | (1) BRAIN | (2) BREAST | (29) CELL-LINE | (1) CERVIX | (3) HELA | (6) LIVER | (1) OTHER | (1) RRP40.ip | (6) SKIN | (1) TESTES | (1) XRN.ip |
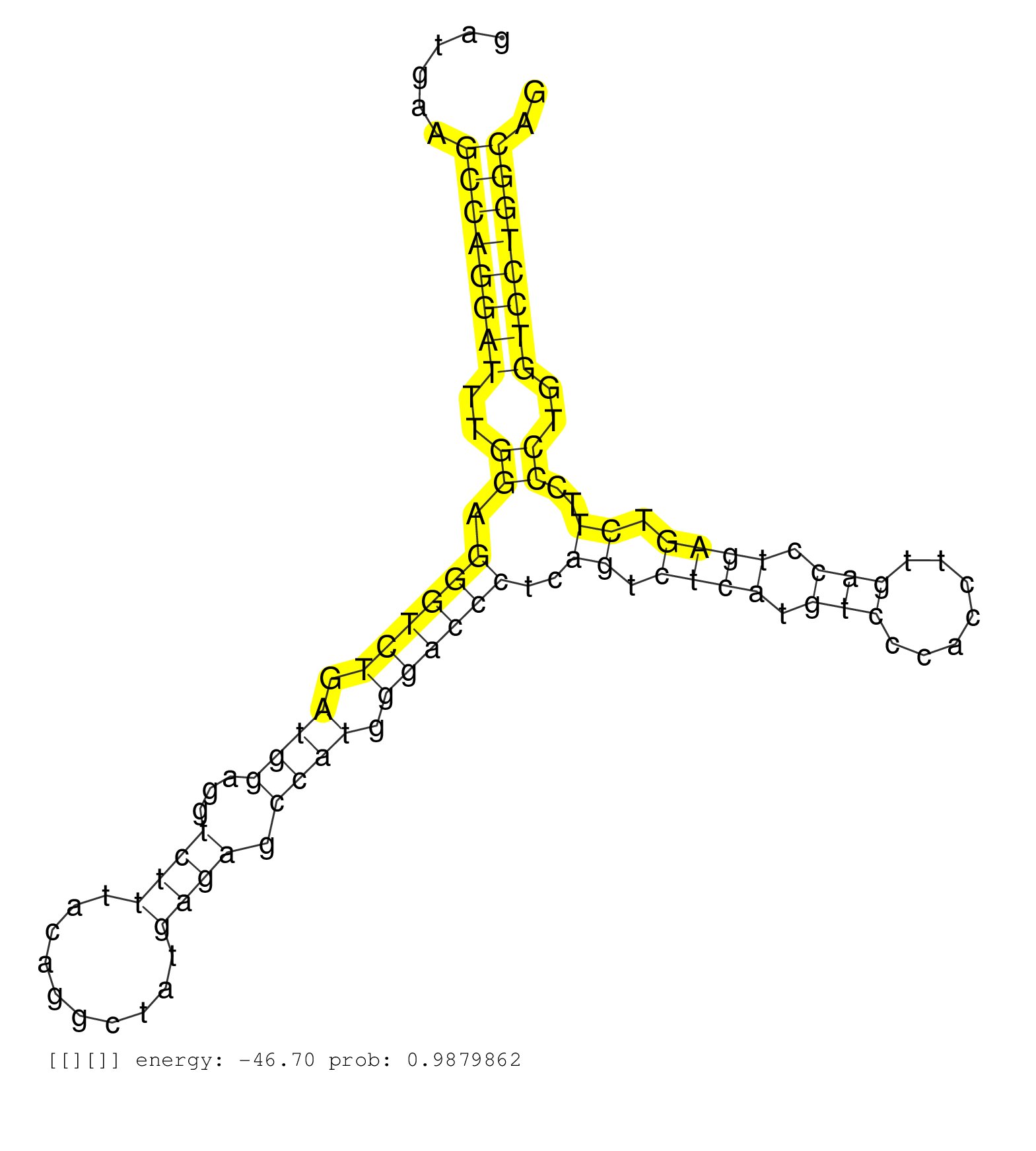
| GTGGGAACAGTGTCGTTAAAGGACAGAACTAAATGTGTGGGGTGGAGGGAAGAGGAGCTGGCTGGAGTTGAGTGGGAGAGGCTGAGTGGAGATGAAGCCAGGATTTGGAGGGTCTGATGGAGGTCTTTACAGGCTATGAGAGCCATGGGACCCTCAGTCTCATGTCCCACCTTGACCTGAGTCTTCCCTGGTCCTGGCAGGTGGAAGTGCAGTGCCTGCTCTTGCTGGAGGTGCTGGGTGGCTCCCACAA ................................................................................................((((((((..((.((((((.((((...((((..........)))).)))).))))))..((.((((.(((.......))).)))).))..))..)))))))).................................................... ..........................................................................................91...........................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR189782 | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR553576(SRX182782) source: Testis. (testes) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR189784 | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR038858(GSM458541) MEL202. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189786 | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191408(GSM715518) 88genomic small RNA (size selected RNA from t. (breast) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR029128(GSM416757) H520. (cell line) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGA..................................................................................................................................... | 22 | 1 | 32.00 | 32.00 | 4.00 | 1.00 | - | 4.00 | 4.00 | - | 2.00 | - | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | 2.00 | 2.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGTCTTCCCTGGTCCTGGCAGA................................................. | 22 | 1 | 12.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCT....................................................................................................................................... | 20 | 1 | 7.00 | 7.00 | - | - | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTG...................................................................................................................................... | 21 | 1 | 6.00 | 6.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAA.................................................................................................................................... | 23 | 1 | 5.00 | 32.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGTCTTCCCTGGTCCTGGCAGAA................................................ | 23 | 1 | 5.00 | 1.00 | - | 2.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAAG................................................................................................................................... | 24 | 1 | 4.00 | 32.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGAGTCTTCCCTGGTCCTGGCAGA................................................. | 24 | 3.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................CCAGGATTTGGAGGGTCTGATGGAGGTCTTTACAGGCTATGAGAGCCATGGGACCCTCAGTCTCATGTC.................................................................................... | 69 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GTGGAGATGAAGCCAGGATTTGGA............................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAAA................................................................................................................................... | 24 | 1 | 2.00 | 32.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGT..................................................................................................................................... | 22 | 1 | 2.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGGGC....................................................................................................................................... | 20 | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAAT................................................................................................................................... | 24 | 1 | 2.00 | 32.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAT.................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................GAGGAGCTGGCTGGAGTTGAGT................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGGCT....................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGATGGA................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGTGGAAGTGCAGTGCCTG................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............TTAAAGGACAGAACTAAATGTGTGGG................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTA...................................................................................................................................... | 21 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTCAAA................................................................................................................................... | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................GTCCTGGCAGGTGGAAGA.......................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................AGTCTTCCCTGGTCCTGGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AAGGACAGAACTAAATGTGTGGGGGGGG............................................................................................................................................................................................................ | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGTGTGA..................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ..................................................AGAGGAGCTGGCTGGTA....................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............TTAAAGGACAGAACTAAATGTGTGG.................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................GGGAAGAGGAGCTGGCTT.......................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .....................................................................................................GATTTGGAGGGTCTGATGGAGGA.............................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGGCCG...................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................AGTCTTCCCTGGTCCTGGCAGAAT............................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GAGGTGCTGGGTGGCGG...... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAAAA.................................................................................................................................. | 25 | 1 | 1.00 | 32.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTGACCTGAGTCTTCCCTGGTCCTGGCAA.................................................. | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................GGGAAGAGGAGCTGGTTGG......................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................AGAGGAGCTGGCTGGCAAA..................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...............................................................................................AGCCAGGATTTGGAGGGTCTGAAGA.................................................................................................................................. | 25 | 1 | 1.00 | 32.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGAGTCTTCCCTGGTCCTGGCAGC................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TGAGTCTTCCCTGGTCCTGGCAGT................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................AGGAGCTGGCTGGAGTTG.................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ....................................................AGGAGCTGGCTGGAGTT..................................................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| GTGGGAACAGTGTCGTTAAAGGACAGAACTAAATGTGTGGGGTGGAGGGAAGAGGAGCTGGCTGGAGTTGAGTGGGAGAGGCTGAGTGGAGATGAAGCCAGGATTTGGAGGGTCTGATGGAGGTCTTTACAGGCTATGAGAGCCATGGGACCCTCAGTCTCATGTCCCACCTTGACCTGAGTCTTCCCTGGTCCTGGCAGGTGGAAGTGCAGTGCCTGCTCTTGCTGGAGGTGCTGGGTGGCTCCCACAA ................................................................................................((((((((..((.((((((.((((...((((..........)))).)))).))))))..((.((((.(((.......))).)))).))..))..)))))))).................................................... ..........................................................................................91...........................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR189782 | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR553576(SRX182782) source: Testis. (testes) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR189784 | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR038858(GSM458541) MEL202. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189786 | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191408(GSM715518) 88genomic small RNA (size selected RNA from t. (breast) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR029128(GSM416757) H520. (cell line) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................TGTCCCACCTTGACCTGTGG.................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................GCCAGGATTTGGAGGGTCTG...................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................CTGGTCCTGGCAGGTCCAG............................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................CCTGGCAGGTGGAAGCCT........................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................GCTGGAGTTGAGTGGGAG............................................................................................................................................................................ | 18 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | 0.17 |