| (1) AGO1.ip | (1) AGO2.ip | (1) BRAIN | (3) BREAST | (12) CELL-LINE | (1) CERVIX | (5) HEART | (2) HELA | (9) LIVER | (1) OTHER | (6) SKIN |
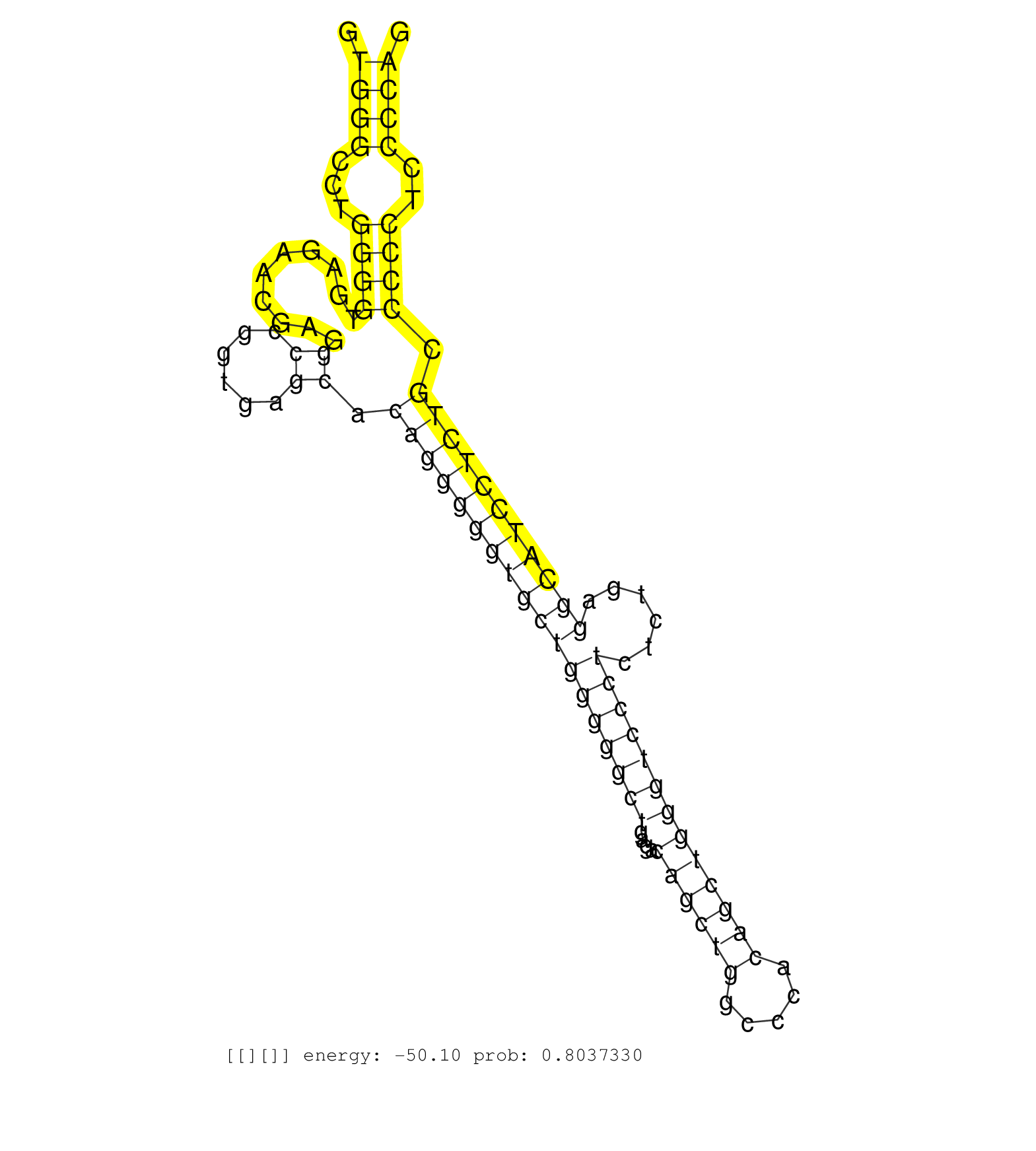
| TCCAGTCCATGCTGCGTCGCTGGTTCGTGGAGACGGAGGGGGCTGTCAAGGTGGGCCTGGGGTGAGAACGAGGCCGGTGAGCACAGGGGGTGCTGGGGGCTGAGACAGCTGGCCCACAGCTGGGTCCCTCTCTGAGGCATCCTCTGCCCCCTCCCCAGGCCTACTTGTGGGACAACAACCAGGTGGTTGTGCAGTGGCTGGAACAGCA ...................................................((((...((((..........((......)).((((((((((((((((((....((((((.....)))))))))))))......))))))))))).))))..))))................................................... ..................................................51.........................................................................................................158................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR553574(SRX182780) source: Heart. (Heart) | SRR038859(GSM458542) MM386. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM532876(GSM532876) G547T. (cervix) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577738(Rovira) total RNA. (breast) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | SRR343335 | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189782 | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGT................................................. | 22 | 1 | 9.00 | 2.00 | - | - | - | 1.00 | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............CGTCGCTGGTTCG..................................................................................................................................................................................... | 13 | 2 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGA................................................. | 22 | 1 | 7.00 | 2.00 | - | 1.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCTGGGGTGAGAACGAG........................................................................................................................................ | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCA................................................... | 20 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GCGTCGCTGGTTCGTTT.................................................................................................................................................................................. | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GTGGAGACGGAGGGGGCTGTCAAG.............................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCTGGGGTGAGAACGAGGCCGG................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................AGGTGGTTGTGCAGTGGCT......... | 19 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AACCAGGTGGTTGTGCAGTG............ | 20 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CAGGTGGTTGTGCAGATAA.......... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................CGGTGAGCACAGGGGG...................................................................................................................... | 16 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | 0.50 | - | - | - |
| .......................................................................GGCCGGTGAGCACAGTAGA...................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................CTGGTTCGTGGAGACGGAGGGGG...................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AACCAGGTGGTTGTGCAGTGGC.......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGCGTCGCTGGTTCGTGGAGACGGAGGGG....................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGGTTGTGCAGTGGCTTC....... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCCTGGGGTGAGAACGA......................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCTGGGGTGAGAACGAGGCCGGTGAGCACAGGGGGTGCTGGGGGCTGAGACAGCTGGCCCACAGCTGGGTCCCTCTCTG.......................................................................... | 84 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGTAT............................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGATA............................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATGCTGCGTCGCTGGTTCGTGGAGAC.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCTGGTTCGTGGAGACGGAG.......................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGAT................................................ | 23 | 1 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ATCCTCTGCCCCCTCCCCAGT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCCTGGGGTGAGAACGAGG....................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACCAGGTGGTTGTGCAGTG............ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGGTTCGTGGAGACGGAG.......................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACAACCAGGTGGTTGGC................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TCCCTCTCTGAGGCATCCTCTGT............................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGATT............................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGTAG............................................... | 24 | 1 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCAGTAA............................................... | 24 | 1 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CATCCTCTGCCCCCTCCCCACA................................................. | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGTGCTGGGGGCTGACT....................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............GTCGCTGGTTCGT.................................................................................................................................................................................... | 13 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..............CGTCGCTGGTTC...................................................................................................................................................................................... | 12 | 8 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CGGTGAGCACAGGGGA...................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - |
| ..........................................................................CGGTGAGCACAGGGG....................................................................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TCGCTGGTTCGTG................................................................................................................................................................................... | 13 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| .....................................................................................................................................................................TGTGGGACAACAACC............................ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| ......................GTTCGTGGAGACG............................................................................................................................................................................. | 13 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| TCCAGTCCATGCTGCGTCGCTGGTTCGTGGAGACGGAGGGGGCTGTCAAGGTGGGCCTGGGGTGAGAACGAGGCCGGTGAGCACAGGGGGTGCTGGGGGCTGAGACAGCTGGCCCACAGCTGGGTCCCTCTCTGAGGCATCCTCTGCCCCCTCCCCAGGCCTACTTGTGGGACAACAACCAGGTGGTTGTGCAGTGGCTGGAACAGCA ...................................................((((...((((..........((......)).((((((((((((((((((....((((((.....)))))))))))))......))))))))))).))))..))))................................................... ..................................................51.........................................................................................................158................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR553574(SRX182780) source: Heart. (Heart) | SRR038859(GSM458542) MM386. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM532876(GSM532876) G547T. (cervix) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577738(Rovira) total RNA. (breast) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | SRR343335 | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189782 | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TCCCTCTCTGAGGCAACCA................................................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CAGGTGGTTGTGCAGTG............ | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GGCCGGTGAGCACAGGGCCGC.................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TCCCTCTCTGAGGCAACAA................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |