| (4) B-CELL | (4) BREAST | (7) CELL-LINE | (2) CERVIX | (2) HEART | (2) HELA | (3) LIVER | (2) OTHER | (18) SKIN | (1) TESTES | (1) UTERUS |
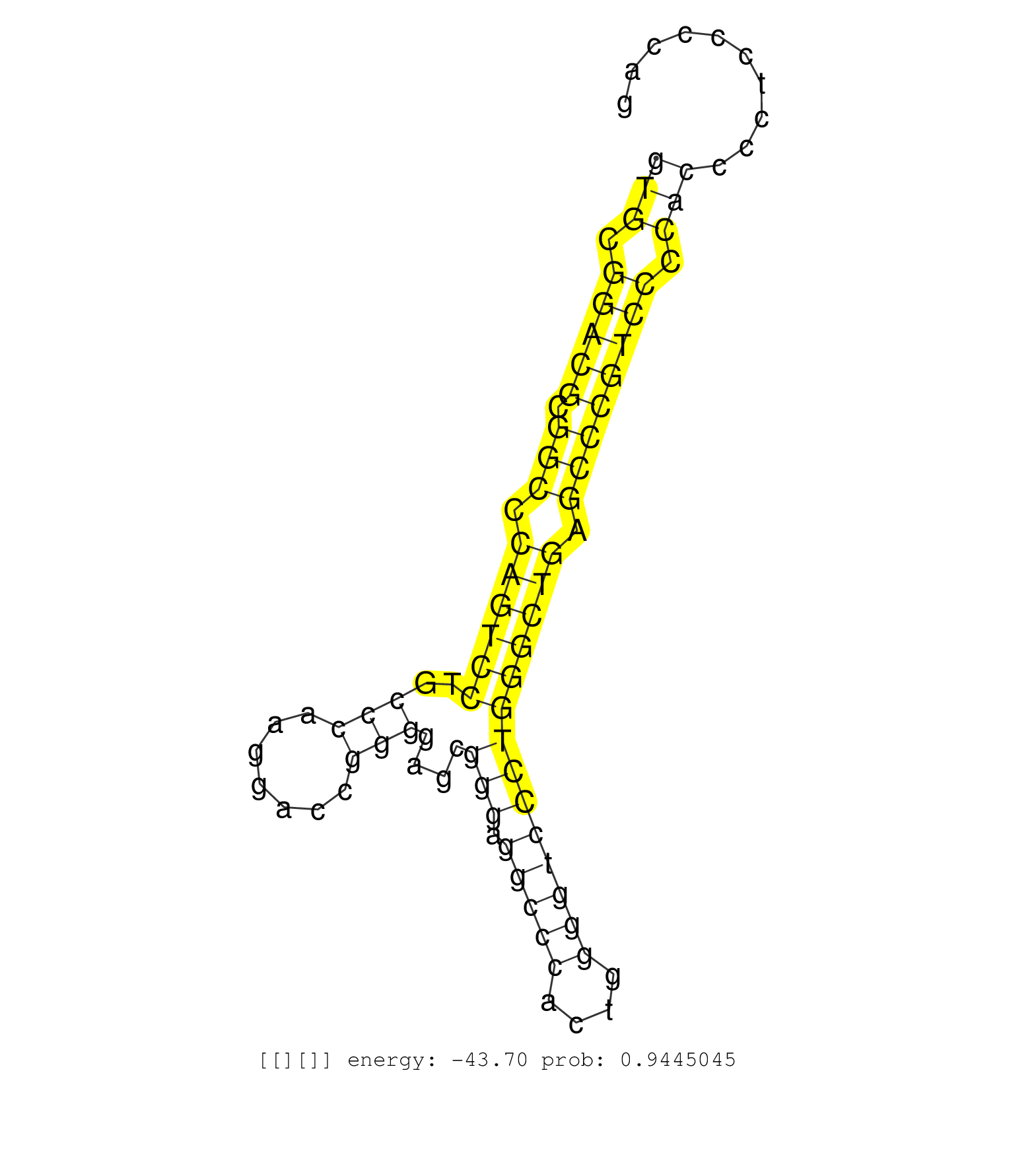
| TGGCTGCCCTCAACAACCACCGCGAGGTGGCCCAGATCCTCATCCGGGAGGTGCGGACGCGGCCCAGTCCTGCCCAAGGACCGGGGAGCGGGAGGCCCACTGGGGTCCCTGGGCTGAGCCCGTCCCCACCCCTCCCCAGGGCCGCTGTGACGTGAACGTGCGCAACCGGAAGCTGCAGTCCCCGCTGCA ..................................................(((.(((((.(((.((((((..(((.......)))....(((.(((((....)))))))))))))).)))))))).)))............................................................ ..................................................51......................................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189782 | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR189784 | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM541797(GSM541797) differentiated human embryonic stem cells. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038858(GSM458541) MEL202. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM956925F181A(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR189787 | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................TGCGGACGCGGCCCAGTCCTG..................................................................................................................... | 21 | 1 | 7.00 | 7.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTCCTGC.................................................................................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTCCTGCC................................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTCCT...................................................................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTCCTGT.................................................................................................................... | 22 | 1 | 3.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CTGGGCTGAGCCCGTCCCCACT........................................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................GCCCGTCCCCACCCCTTGC..................................................... | 19 | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................TGTGACGTGAACGTGCGCAACCGGAAGCT............... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGCGGACGCGGCCCAGTCCTG..................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCCTT............................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................CGGGAGGTGCGGACGCGGCCCA........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CGCAACCGGAAGCTGCAGTCCCC...... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................CCGGGGAGCGGGAGGCCCCGGG....................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................CCACTGGGGTCCCTGGGCTGAGCCC.................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCCAC............................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCCACC........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GGTCCCTGGGCTGAGCATC................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCC.............................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCCACCCCTCCCCAGGGCACCC........................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................AGTCCTGCCCAAGGACCGGGGAG..................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CCAAGGACCGGGGAGGTAG................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCCTTT........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTC........................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CCTGGGCTGAGCCCGTCCCCA............................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CCCAGATCCTCATCCGGGAGGG......................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................CTGTGACGTGAACGTGCGCAACCGGAAGC................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGCGGACGCGGCCCAGTCCTGA.................................................................................................................... | 22 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCCACCCCTCCCCAGGGCCGC............................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGTGACGTGAACGTGCGCAACCGGAAGCTGC............. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................ACCGGAAGCTGCAGTC......... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AACCGGAAGCTGCAG........... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CCGCGAGGTGGCCC............................................................................................................................................................ | 14 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| TGGCTGCCCTCAACAACCACCGCGAGGTGGCCCAGATCCTCATCCGGGAGGTGCGGACGCGGCCCAGTCCTGCCCAAGGACCGGGGAGCGGGAGGCCCACTGGGGTCCCTGGGCTGAGCCCGTCCCCACCCCTCCCCAGGGCCGCTGTGACGTGAACGTGCGCAACCGGAAGCTGCAGTCCCCGCTGCA ..................................................(((.(((((.(((.((((((..(((.......)))....(((.(((((....)))))))))))))).)))))))).)))............................................................ ..................................................51......................................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189782 | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR189784 | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM541797(GSM541797) differentiated human embryonic stem cells. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038858(GSM458541) MEL202. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM956925F181A(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR189787 | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................CGGCCCAGTCCTGCCCA................................................................................................................. | 17 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CCGTCCCCACCCCTCCCCA................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....TGCCCTCAACAACCATAGG...................................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................GCGGCCCAGTCCTGCCCAAGGA............................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CCGTCCCCACCCCTCCCCAT.................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................CCGTCCCCACCCCTCCAGGT.................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................GCTGCAGTCCCCGCTGGAC | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................CCCGTCCCCACCCCTCCC..................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - |
| ..........................................................GCGGCCCAGTCCTGC.................................................................................................................... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ............................................................................................................................................................CGTGCGCAACCG..................... | 12 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| .................................AGATCCTCATCCGG.............................................................................................................................................. | 14 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - |
| ...................................................................................................................................CTCCCCAGGGCCGCT........................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |