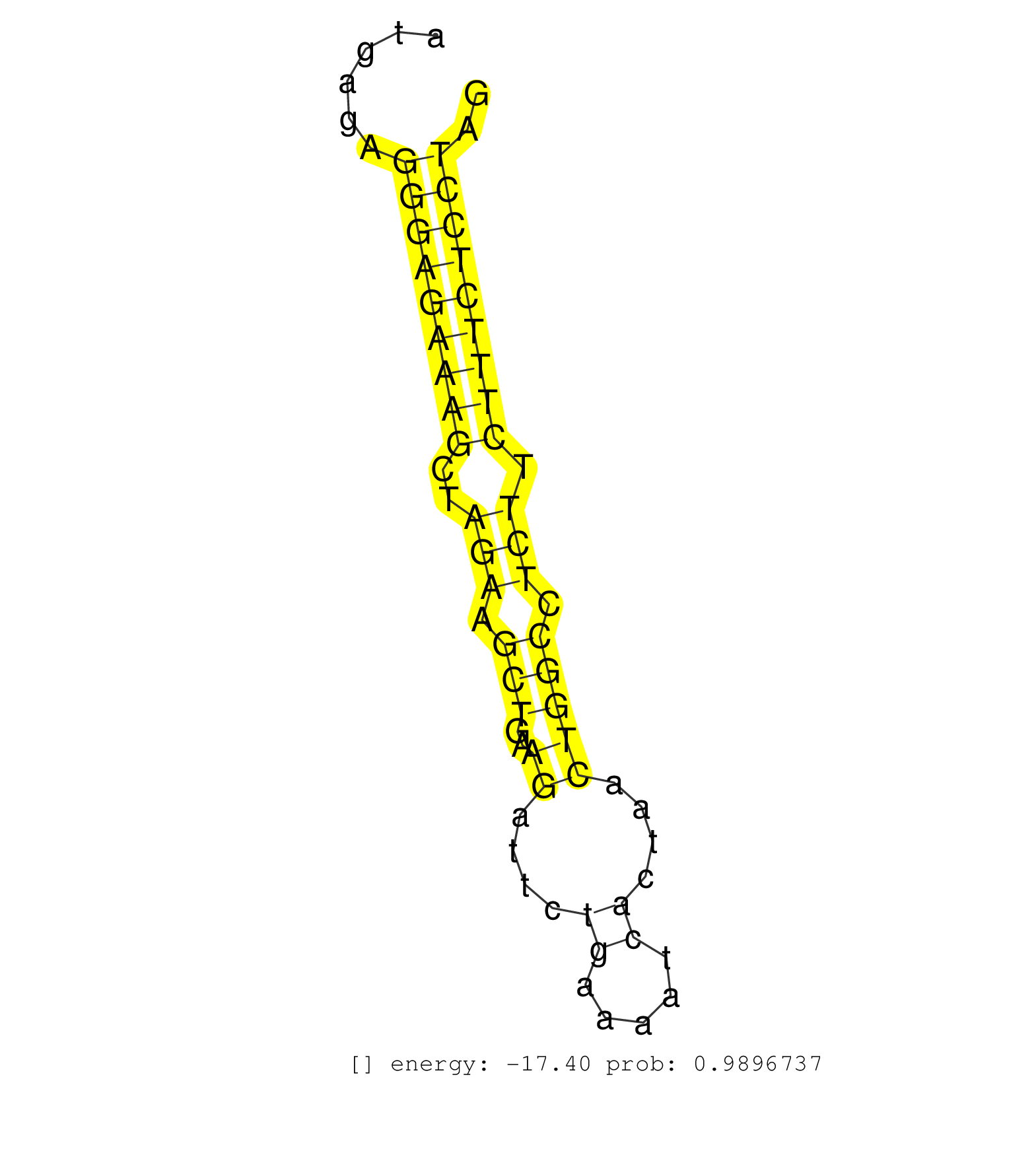
| (2) AGO1.ip | (4) AGO2.ip | (15) B-CELL | (13) BRAIN | (20) BREAST | (29) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (4) HEART | (2) HELA | (1) KIDNEY | (11) LIVER | (3) OTHER | (14) SKIN | (3) UTERUS | (1) XRN.ip |
| ACCTTGGCCTCCCAAAGTTCTGGGATTATAGACATGAGCCACCGCGCCCTGCCAGGGAATTGATTTTTATCTGCCATTGCCCTCTCTCAGTAGGACAGTTCCTTTCTCTTCACCTCTTCTGTCAGCTAAAGACCATGAGAGGGAGAAAGCTAGAAGCTGAAGATTCTGAAAATCACTAACTGGCCTCTTCTTTCTCCTAGGTGTGGCACATTGGTTGACCTTTGCCAGGGCCCCCACCTTCGGCATACTG ............................................................................................................................................(((((((((..(((.(((..((....((.....))....))))).))).))))))))).................................................... ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR038863(GSM458546) MM603. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029131(GSM416760) MCF7. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR390723(GSM850202) total small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR191528(GSM715638) 130genomic small RNA (size selected RNA from . (breast) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR037937(GSM510475) 293cand2. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | TAX577579(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037931(GSM510469) 293GFP. (cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR038856(GSM458539) D11. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191461(GSM715571) 35genomic small RNA (size selected RNA from t. (breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191627(GSM715737) 45genomic small RNA (size selected RNA from t. (breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577742(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR040036(GSM532921) G243N. (cervix) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | TAX577746(Rovira) total RNA. (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | TAX577740(Rovira) total RNA. (breast) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAG........................................................................................ | 23 | 1 | 54.00 | 54.00 | 2.00 | 6.00 | - | 1.00 | 4.00 | - | 4.00 | - | 1.00 | 1.00 | - | 2.00 | 4.00 | - | - | - | 4.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 2.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | - | 2.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGA.......................................................................................... | 21 | 1 | 38.00 | 38.00 | - | - | 4.00 | 2.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | 3.00 | - | - | - | - | 2.00 | 1.00 | 3.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGAT...................................................................................... | 25 | 1 | 16.00 | 16.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAGT................................................. | 22 | 1 | 14.00 | 5.00 | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCT............................................................................................ | 19 | 1 | 14.00 | 14.00 | - | 1.00 | - | - | - | - | - | 4.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGA....................................................................................... | 24 | 1 | 12.00 | 12.00 | - | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAA......................................................................................... | 22 | 1 | 10.00 | 10.00 | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCTGAAGATT..................................................................................... | 27 | 1 | 6.00 | 6.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCTGA.......................................................................................... | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTG........................................................................................... | 20 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGGAGAAAGCTAGAAGCTGAAG........................................................................................ | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAG.................................................. | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGT....................................................................................... | 24 | 1 | 3.00 | 54.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAA........................................................................................ | 23 | 1 | 3.00 | 10.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGATT..................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCT............................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCTG........................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGGCCTCCCAAAGTTCTCGTT................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................TGAGCCACCGCGCCCTGCCAGACTC............................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAGTA................................................ | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AAGATTCTGAAAATCACTAACTGGCCTCTT............................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAGA................................................. | 22 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGC............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGGAGAAAGCTAGAAGCTGAAGATTCTGAAAA.............................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGGAGAAAGCTAGAAGCTGAAGA....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGCAG........................................................................................ | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAAAA................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................TGGGATTATAGACATGAGCCA................................................................................................................................................................................................................. | 21 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGAAGCTGAAGATTCG................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGCAGA....................................................................................... | 24 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGACG........................................................................................ | 23 | 1 | 1.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCTGAAGAT...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTTA.......................................................................................... | 21 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CTAACTGGCCTCTTCTTTCTCCTAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CATGAGCCACCGCGCCCCAAT..................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTATT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................TGGTTGACCTTTGCCAGGGC................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................CTTTCTCCTAGGTGTGGCACATT...................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCGGAA......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAGTT................................................ | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGC....................................................................................... | 24 | 1 | 1.00 | 54.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGGGATTATAGACATGAGCTCTG............................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGG....................................................................................... | 24 | 1 | 1.00 | 54.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAAA....................................................................................... | 24 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................CCCACCTTCGGCATACTAAAC | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................................................................................................................................GAGGGAGAAAGCTAGAAGCTGAA......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGGAGAAAGCTAGAAGCTG........................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CATGAGAGGGAGAAATTT................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGT.......................................................................................... | 21 | 1 | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGCTGAAGTAA..................................................................................... | 26 | 1 | 1.00 | 54.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGGGAGAAAGCTAGAAGTTGA.......................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................GGTTGACCTTTGCCAGGGCCCCC............... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGGCCTCTTCTTTCTCCTAATT................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................ATTGGTTGACCTTTGC......................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| .....................GGGATTATAGACATGAGCCACCGCGCCC......................................................................................................................................................................................................... | 28 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACCTTGGCCTCCCAAAGTTCTGGGATTATAGACATGAGCCACCGCGCCCTGCCAGGGAATTGATTTTTATCTGCCATTGCCCTCTCTCAGTAGGACAGTTCCTTTCTCTTCACCTCTTCTGTCAGCTAAAGACCATGAGAGGGAGAAAGCTAGAAGCTGAAGATTCTGAAAATCACTAACTGGCCTCTTCTTTCTCCTAGGTGTGGCACATTGGTTGACCTTTGCCAGGGCCCCCACCTTCGGCATACTG ............................................................................................................................................(((((((((..(((.(((..((....((.....))....))))).))).))))))))).................................................... ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR038863(GSM458546) MM603. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029131(GSM416760) MCF7. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR390723(GSM850202) total small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR191528(GSM715638) 130genomic small RNA (size selected RNA from . (breast) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR037937(GSM510475) 293cand2. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | TAX577579(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037931(GSM510469) 293GFP. (cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR038856(GSM458539) D11. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191461(GSM715571) 35genomic small RNA (size selected RNA from t. (breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191627(GSM715737) 45genomic small RNA (size selected RNA from t. (breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577742(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR040036(GSM532921) G243N. (cervix) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | TAX577746(Rovira) total RNA. (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | TAX577740(Rovira) total RNA. (breast) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................GCCACCGCGCCCTGCGCG................................................................................................................................................................................................... | 18 | 6.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................GCCTCTTCTTTCTCCTTGAC................................................ | 20 | 5.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................GCCACCGCGCCCTGCCATG.................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................GCCTCTTCTTTCTCCTTGAG................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................GCCTCTTCTTTCTCCTTGA................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................GGATTATAGACATGAGCCACCTATC........................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..CTTGGCCTCCCAAAGTTCTGGGATAT.............................................................................................................................................................................................................................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .CCTTGGCCTCCCAAAGTTCTGGAG................................................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................CTTCTGTCAGCTAAAGT...................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................GAGCCACCGCGCCCTGGTGA................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................GGGCCCCCACCTTCGG....... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ..........................................................................CATTGCCCTCTCTCAG................................................................................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |