| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (1) AGO3.ip | (1) B-CELL | (4) BRAIN | (10) BREAST | (15) CELL-LINE | (1) CERVIX | (2) FIBROBLAST | (8) HEART | (4) HELA | (1) LIVER | (1) OTHER | (12) SKIN | (1) UTERUS | (1) XRN.ip |
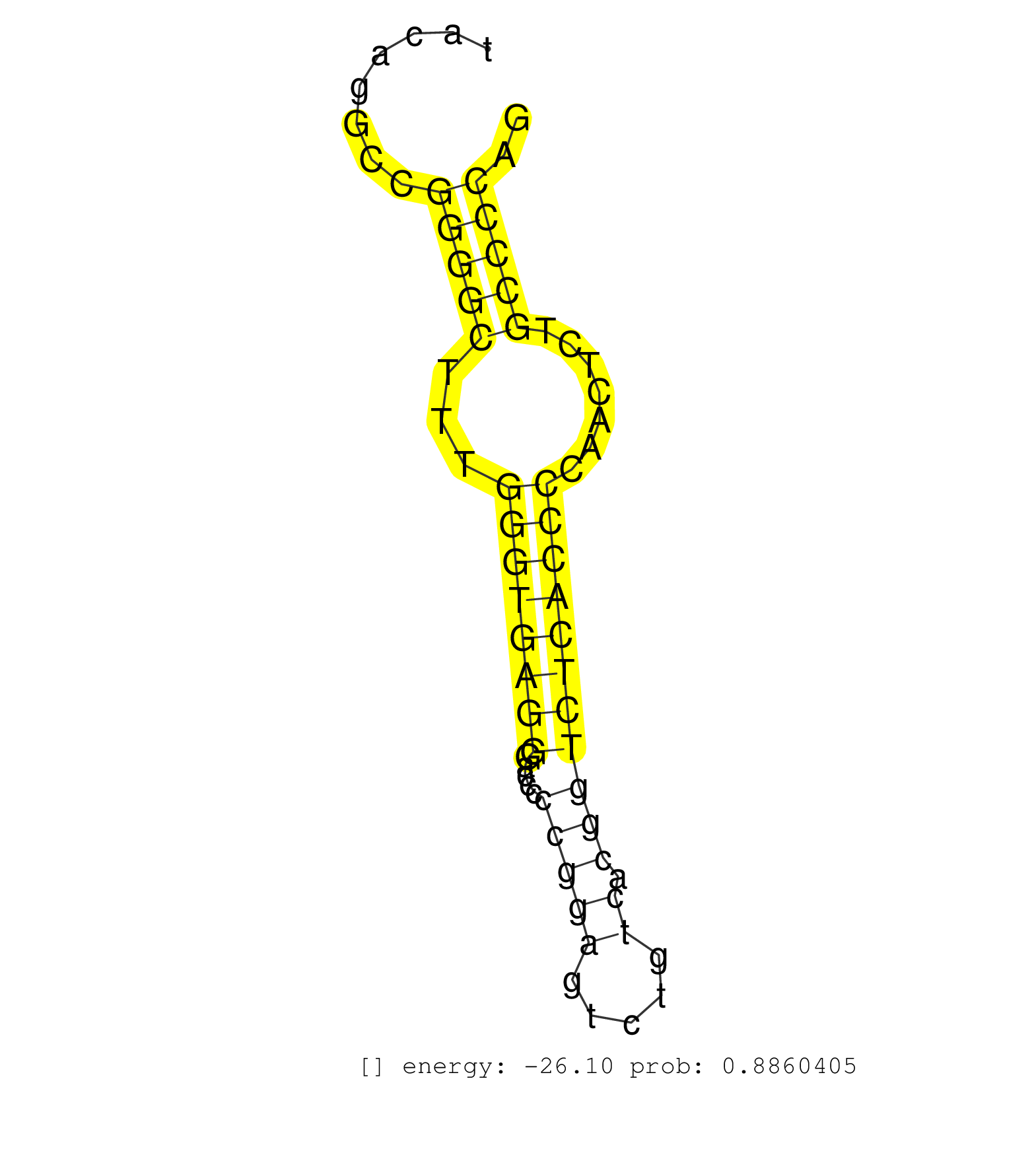
| GGGCCAGGCTGTGTGGCTGGAGGGGGTCCCTGCAGCCTCCTCAGTGAGCTCCCCTCTCACTCTAGGTCAGAAGAGAGTGAGGAGCGGGGGCAGGGTGACCTGGGGACGGGCTTGGGCTCTGTTCCCTGGAGGTTACAGGCCGGGGCTTTGGGTGAGGGACCCCCGGAGTCTGTCACGGTCTCACCCCAACTCTGCCCCAGGGAACAGCTTTGTCAGGCACAATGCCCGCCAGCTGACCCGCGTGTACCCG .............................................................................................................................................(((((...((((((((.....(((((.....)).))))))))))).......))))).................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191460(GSM715570) 33genomic small RNA (size selected RNA from t. (breast) | SRR191567(GSM715677) 46genomic small RNA (size selected RNA from t. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR189784 | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR189782 | SRR038863(GSM458546) MM603. (cell line) | SRR191593(GSM715703) 62genomic small RNA (size selected RNA from t. (breast) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191513(GSM715623) 164genomic small RNA (size selected RNA from . (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189785 | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR029131(GSM416760) MCF7. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAGT................................................. | 23 | 1 | 35.00 | 6.00 | 3.00 | 6.00 | 4.00 | 3.00 | 1.00 | - | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................GTCTCACCCCAACTCTGCCCCAG.................................................. | 23 | 1 | 14.00 | 14.00 | 4.00 | - | - | - | 1.00 | - | - | - | 3.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................GTCTCACCCCAACTCTGCCCCAGT................................................. | 24 | 1 | 10.00 | 14.00 | - | - | - | - | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAGA................................................. | 23 | 1 | 8.00 | 6.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAG.................................................. | 22 | 1 | 6.00 | 6.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................GCCGGGGCTTTGGGTGAGGG............................................................................................ | 20 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGG............................................................................................. | 18 | 1 | 5.00 | 5.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................CGGGGCTTTGGGTGAGGGAC.......................................................................................... | 20 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAAT................................................. | 23 | 1 | 4.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGGGACC......................................................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAGC................................................. | 23 | 1 | 3.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGGGAC.......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAGTT................................................ | 24 | 1 | 2.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAGAT................................................ | 24 | 1 | 2.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CGGGGCTTTGGGTGAGGG............................................................................................ | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAAAA................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCACCCCAACTCTGCCCCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................CTTGGGCTCTGTTCCCTGGAGGTTACAGGCCGGG.......................................................................................................... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGGAACAGCTTTGTCAG.................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGGG............................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CGGGGCTTTGGGTGAGTGAC.......................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GTCTCACCCCAACTCTGCCCCAT.................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GAGGTTACAGGCCGGGATC....................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........TGTGGCTGGAGGGGGCTTG............................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCC................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCATT................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGGGACA......................................................................................... | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................AGGGTGACCTGGGGAAGGC............................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CGGGGCTTTGGGTGAGGGACCC........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAAA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCACC................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................CGGAGTCTGTCACGG........................................................................ | 15 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAA.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GCCAGGCTGTGTGGCTGGAGG................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCACCCCAACTCTGCCCCAATA................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCGGGGCTTTGGGTGAGGGACCCA....................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................GGGGACGGGCTTGGGAGAG.................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................GGAGGTTACAGGCCGGGTGGC...................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CGGGGCTTTGGGTGAGG............................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GGGGCTTTGGGTGAGG............................................................................................. | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| GGGCCAGGCTGTGTGGCTGGAGGGGGTCCCTGCAGCCTCCTCAGTGAGCTCCCCTCTCACTCTAGGTCAGAAGAGAGTGAGGAGCGGGGGCAGGGTGACCTGGGGACGGGCTTGGGCTCTGTTCCCTGGAGGTTACAGGCCGGGGCTTTGGGTGAGGGACCCCCGGAGTCTGTCACGGTCTCACCCCAACTCTGCCCCAGGGAACAGCTTTGTCAGGCACAATGCCCGCCAGCTGACCCGCGTGTACCCG .............................................................................................................................................(((((...((((((((.....(((((.....)).))))))))))).......))))).................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191460(GSM715570) 33genomic small RNA (size selected RNA from t. (breast) | SRR191567(GSM715677) 46genomic small RNA (size selected RNA from t. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR189784 | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR189782 | SRR038863(GSM458546) MM603. (cell line) | SRR191593(GSM715703) 62genomic small RNA (size selected RNA from t. (breast) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191513(GSM715623) 164genomic small RNA (size selected RNA from . (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189785 | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR029131(GSM416760) MCF7. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................ACAGGCCGGGGCTTTG.................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTTGGGCTCTGTTCCAGAC......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................AGGAGCGGGGGCAGGGTC......................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....AGGCTGTGTGGCTGGAGCAGT................................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................GTCCCTGCAGCCTCCGACT.............................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .....AGGCTGTGTGGCTGGAGCAGC................................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |