| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (2) AGO3.ip | (13) B-CELL | (7) BREAST | (45) CELL-LINE | (3) CERVIX | (2) FIBROBLAST | (1) HEART | (4) HELA | (1) KIDNEY | (5) LIVER | (2) OTHER | (1) RRP40.ip | (7) SKIN | (2) UTERUS | (1) XRN.ip |
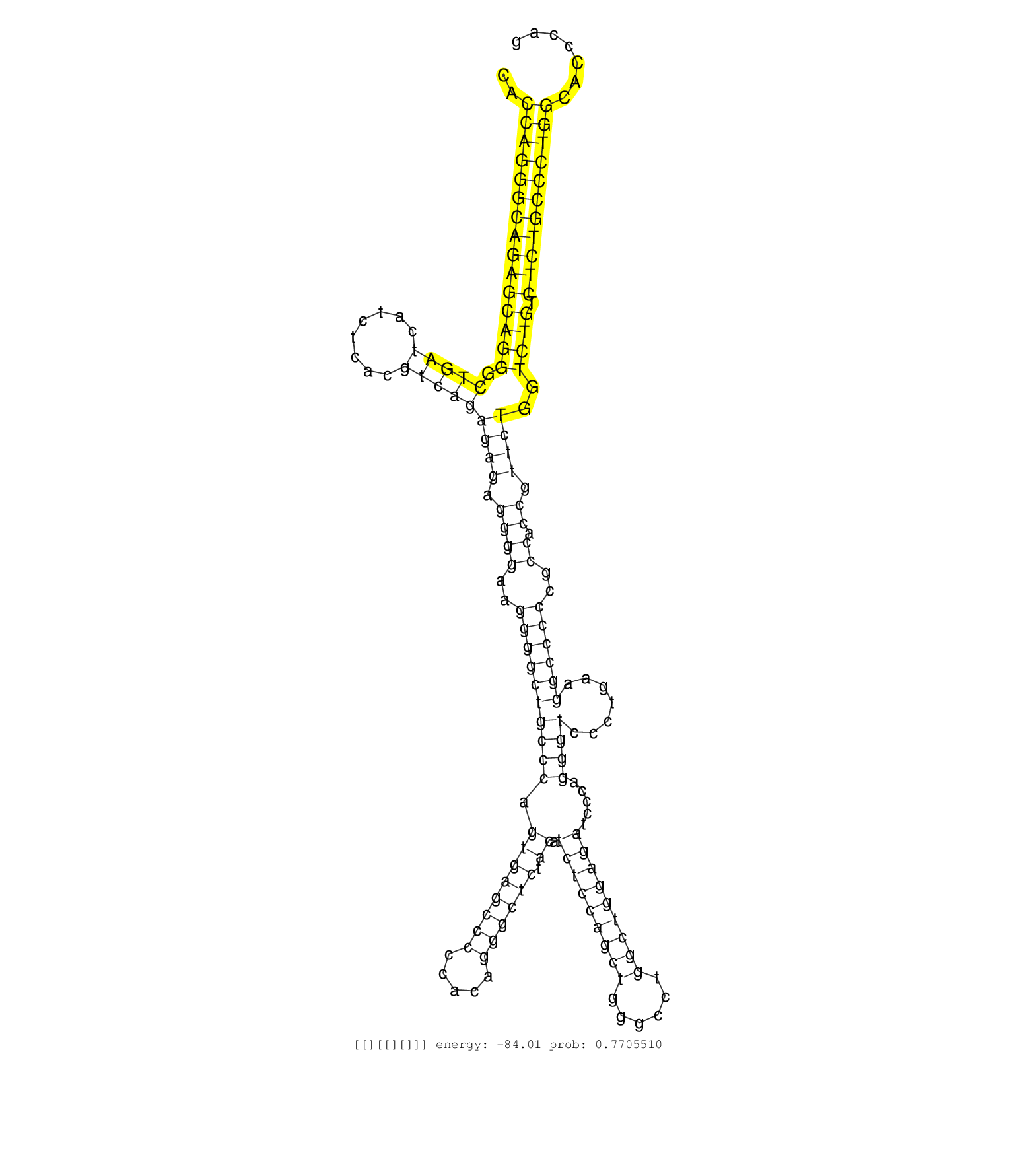
| AAGCAATTGCCAATCCATGTGGTGTCTTTGGGCCCCCACCAGGGCAGAGCAGGGCTGATCATCTCACGTCAGAGAGAGGGGAAGGGGCTGCCCAGTGAGCCCCCACAGGGCTCTACATCTCCAGCTGGGCCTGGCTGGAGATCCCAGGGTCCCTGAAGGCCCCCGCCACCGTTCTGGTCTGTCTCTGCCCTGGCACCCAGATGGAGGAGGGAGTGAGTGCCGTGGCTCACCGCATCCCCTGCCGGGACAT ......................................(((((((((((((((.(((((........)))))((((.((((..((((((((((.((((((((.....)))))).)).(((((((((......))))))))).....)))).......))))))..)).)).))))..)))).)))))))))))......................................................... ....................................37.................................................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR029130(GSM416759) DLD2. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR189786 | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR015358(GSM380323) Na╠°ve B Cell (Naive39). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577453(Rovira) total RNA. (breast) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR040028(GSM532913) G026N. (cervix) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR390723(GSM850202) total small RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029131(GSM416760) MCF7. (cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040027(GSM532912) G220T. (cervix) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAT...................................................... | 22 | 1 | 70.00 | 24.00 | 17.00 | 3.00 | 2.00 | 2.00 | - | 2.00 | - | - | 2.00 | - | 1.00 | 3.00 | 1.00 | - | 3.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | 1.00 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 2.00 | - | 1.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAC...................................................... | 22 | 1 | 29.00 | 29.00 | 1.00 | 1.00 | 3.00 | - | - | - | 1.00 | 4.00 | - | 3.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 3.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCA....................................................... | 21 | 1 | 24.00 | 24.00 | 3.00 | 4.00 | - | - | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATA..................................................... | 23 | 1 | 14.00 | 24.00 | 1.00 | 1.00 | - | - | 8.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGA................................................................................................................................................................................................ | 22 | 1 | 12.00 | 12.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGC........................................................ | 20 | 1 | 12.00 | 12.00 | - | - | - | - | - | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATT..................................................... | 23 | 1 | 10.00 | 24.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAA...................................................... | 22 | 1 | 9.00 | 24.00 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACT..................................................... | 23 | 1 | 7.00 | 29.00 | - | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAT............................................................................................................................................................................................... | 23 | 1 | 6.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAA............................................................................................................................................................................................... | 23 | 1 | 6.00 | 12.00 | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAAA.............................................................................................................................................................................................. | 24 | 1 | 5.00 | 12.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CCACCAGGGCAGAGCAGGGCTGAT............................................................................................................................................................................................... | 24 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAAT..................................................... | 23 | 1 | 4.00 | 24.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACA..................................................... | 23 | 1 | 3.00 | 29.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAAA..................................................... | 23 | 1 | 3.00 | 24.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAGA.............................................................................................................................................................................................. | 24 | 1 | 3.00 | 12.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAAT.............................................................................................................................................................................................. | 24 | 1 | 3.00 | 12.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACC..................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CCACCAGGGCAGAGCAGGGCTGAA............................................................................................................................................................................................... | 24 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................CACCAGGGCAGAGCAGGGCTGATA.............................................................................................................................................................................................. | 24 | 1 | 2.00 | 6.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACCAGGGCAGAGCAGGGCTGAAA.............................................................................................................................................................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCTTT..................................................... | 23 | 1 | 2.00 | 12.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAAAT............................................................................................................................................................................................. | 25 | 1 | 2.00 | 12.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGAAAA............................................................................................................................................................................................. | 25 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCAATAT................................................... | 25 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATC..................................................... | 23 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACTAT................................................... | 25 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTTGC........................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACCA.................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CTGGTCTGTCTCTGCCCTGGCAA...................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................CACCAGGGCAGAGCAGGGCTGATTT............................................................................................................................................................................................. | 25 | 1 | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGG.................................................................................................................................................................................................... | 18 | 3 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCTT...................................................... | 22 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATTT.................................................... | 24 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACCAGGGCAGAGCAGGGCTGAAT.............................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..............................................GAGCAGGGCTGATCATCTCACAAA.................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCCA...................................................... | 22 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGAAA...................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................CCACCAGGGCAGAGCAGGGCTTTAA.............................................................................................................................................................................................. | 25 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATTA.................................................... | 24 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGT........................................................ | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATGAT................................................... | 25 | 1 | 1.00 | 24.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGCC........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACCAGGGCAGAGCAGGGCTGATAT............................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACTA.................................................... | 24 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CTCACGTCAGAGAGAGG........................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATAA.................................................... | 24 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACCAGGGCAGAGCAGGGCTGT................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGCTCACCGCATCCCCCACG....... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................CCCTGAAGGCCCCCGATC.................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATTAT................................................... | 25 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGGGGAAGGGGCTGCCCAGT.......................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGCTCACCGCATCCCAACA........ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................CACCAGGGCAGAGCAGGGC................................................................................................................................................................................................... | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GGTCTGTCTCTGCCCTGGCAT...................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................CACCAGGGCAGAGCAGGGCTG................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CCACCAGGGCAGAGCAGGGCTGATGA............................................................................................................................................................................................. | 26 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCATAGA................................................... | 25 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACTT.................................................... | 24 | 1 | 1.00 | 29.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACAAA................................................... | 25 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACCAGGGCAGAGCAGGGCTGAT............................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCTATT.................................................... | 24 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTGGCACAAT................................................... | 25 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACCAGGGCAGAGCAGGC.................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGTCTGTCTCTGCCCTG.......................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CCACCAGGGCAGAGCAG...................................................................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ....................................CACCAGGGCAGAGCAGGGATGA................................................................................................................................................................................................ | 22 | 3 | 0.33 | 1.00 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CTCACCGCATCCCCTGC........ | 17 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| AAGCAATTGCCAATCCATGTGGTGTCTTTGGGCCCCCACCAGGGCAGAGCAGGGCTGATCATCTCACGTCAGAGAGAGGGGAAGGGGCTGCCCAGTGAGCCCCCACAGGGCTCTACATCTCCAGCTGGGCCTGGCTGGAGATCCCAGGGTCCCTGAAGGCCCCCGCCACCGTTCTGGTCTGTCTCTGCCCTGGCACCCAGATGGAGGAGGGAGTGAGTGCCGTGGCTCACCGCATCCCCTGCCGGGACAT ......................................(((((((((((((((.(((((........)))))((((.((((..((((((((((.((((((((.....)))))).)).(((((((((......))))))))).....)))).......))))))..)).)).))))..)))).)))))))))))......................................................... ....................................37.................................................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR029130(GSM416759) DLD2. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR189786 | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR015358(GSM380323) Na╠°ve B Cell (Naive39). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577453(Rovira) total RNA. (breast) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR040028(GSM532913) G026N. (cervix) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR390723(GSM850202) total small RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029131(GSM416760) MCF7. (cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040027(GSM532912) G220T. (cervix) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................ACCAGGGCAGAGCAGGACGA................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................GCCCAGTGAGCCCCCGC................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................CCAGTGAGCCCCCACAGGTAGA......................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................GCTCTACATCTCCAGTCTG.......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................ACATCTCCAGCTGGGCCTGGCTTGC............................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................CACAGGGCTCTACATCTTT................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................CAGGGCTCTACATCTCCAAGC............................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |