| (4) OTHER |
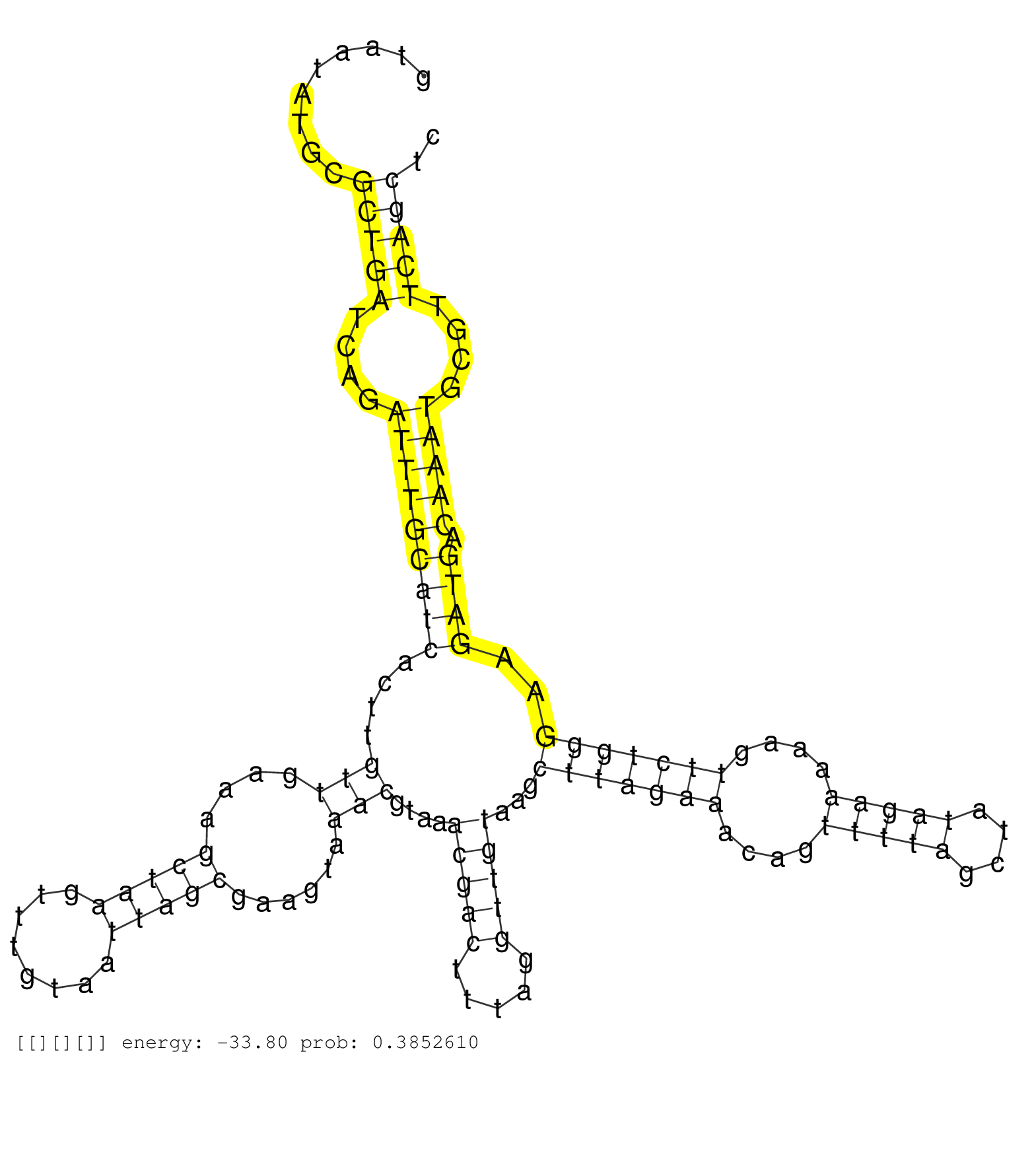
| GCCCCATCAGAACATCAAGCAGTACTTCCAGGAGGCCTACGACTTTATCGGTGAGTATCCCTCATGGTTTAATCCGTACGCATTACCATCCCAACTATGTATACTCATATACTATGTGTATCCCAAAGATACAAGTAATATGCGCTGATCAGATTTGCATCACTTGTTGAAAGCTAAGTTTGTAATTAGCGAAGTAAACGTAAACGACTTTAGGTTGTAAGCTTAGAAACAGTTTTAGCTATAGAAAAAGTTCTGGGAAGATGACAAATGCGTTCAGCTCATTAGGGAAAACGTCTGGATGGTCGTGAAACTTTCCGAATATTTGGCAGAGATTAGGCGGCGTGTCAACC ...............................................................................................................................................(((((....(((((((((....(((....(((((........)))))......)))....(((((.....)))))...(((((((....(((((....)))))....)))))))..)))).)))))....)))))........................................................................ ......................................................................................................................................135..............................................................................................................................................280.................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | V107 | V108 | V040 | V060 |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................GAAGATGACAAATGCGTTCA.......................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................................CATTAGGGAAAACGTCTGGAT.................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ................................................................................................................................................................................AGTTTGTAATTAGCGAAGTAAACGTA.................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ...........................................................................................................................................ATGCGCTGATCAGATTTGC................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................ATCCCAAAGATACAAGTAATATGCGCT............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................................................................................................CGTCTGGATGGTCGTGAAAC....................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................................AGCTCATTAGGGAAAACGTCTGGATGG................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ......................................................................................................................................................................................................................................................................GACAAATGCGTTCAGCTCATTAGGGAA............................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................TGTAAGCTTAGAAACAGTTTTAGCT.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................................................................................TAGGGAAAACGTCTGGATGGT............................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ......................................................................................................................................................AGATTTGCATCACTTGTTGA.................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| GCCCCATCAGAACATCAAGCAGTACTTCCAGGAGGCCTACGACTTTATCGGTGAGTATCCCTCATGGTTTAATCCGTACGCATTACCATCCCAACTATGTATACTCATATACTATGTGTATCCCAAAGATACAAGTAATATGCGCTGATCAGATTTGCATCACTTGTTGAAAGCTAAGTTTGTAATTAGCGAAGTAAACGTAAACGACTTTAGGTTGTAAGCTTAGAAACAGTTTTAGCTATAGAAAAAGTTCTGGGAAGATGACAAATGCGTTCAGCTCATTAGGGAAAACGTCTGGATGGTCGTGAAACTTTCCGAATATTTGGCAGAGATTAGGCGGCGTGTCAACC ...............................................................................................................................................(((((....(((((((((....(((....(((((........)))))......)))....(((((.....)))))...(((((((....(((((....)))))....)))))))..)))).)))))....)))))........................................................................ ......................................................................................................................................135..............................................................................................................................................280.................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | V107 | V108 | V040 | V060 |
|---|---|---|---|---|---|---|---|---|
| .................................................................aactCGTACGGATTATCAA.......................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | |
| .................................................................atctCGTACGGATTATCTA.......................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - |