| (3) OTHER |
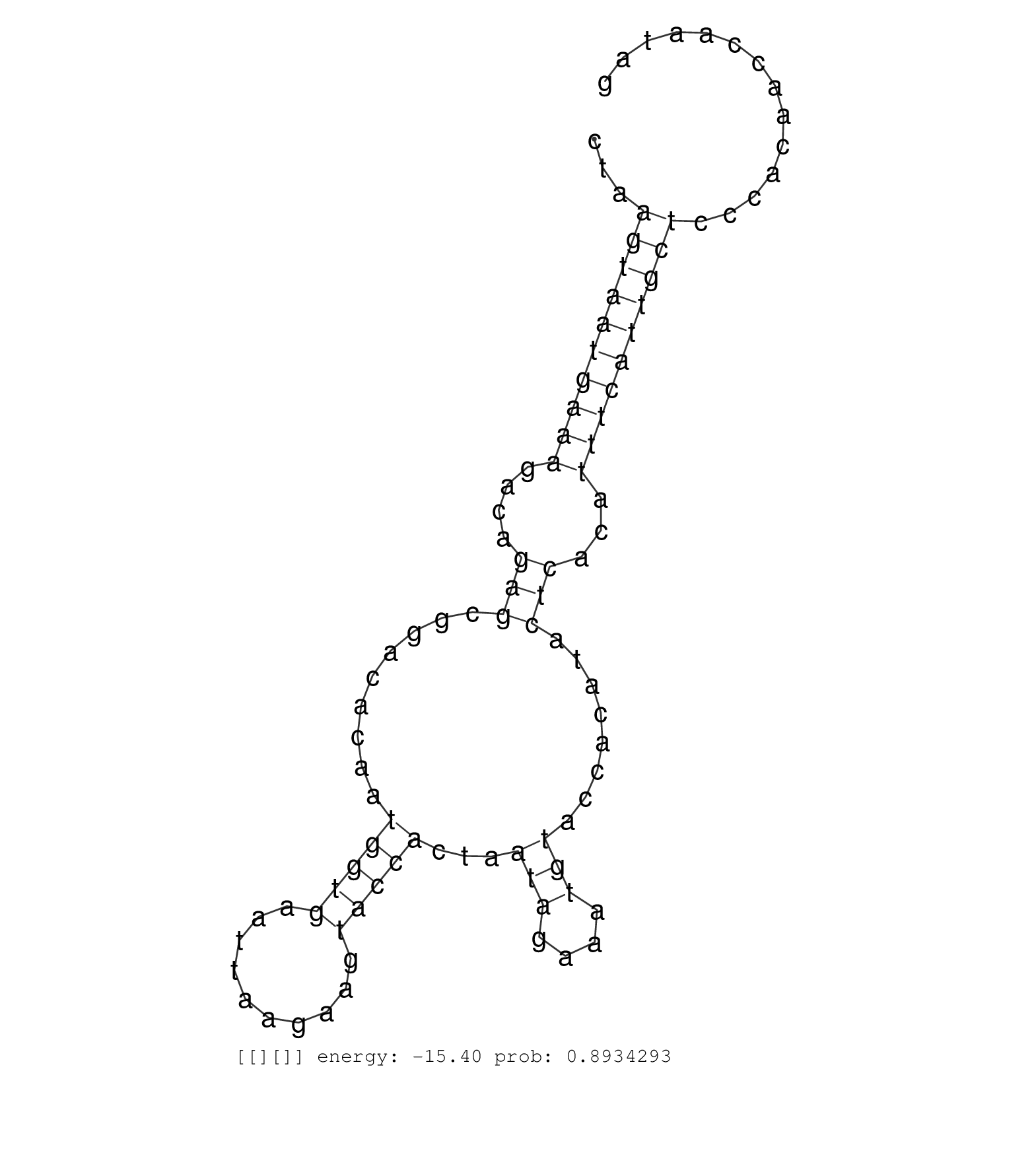
| GATCAAACTGGCGCAACATTCGTTGTCCTGTCTCTTCGGCACGTTCCTATGTAATTCGCTCAGAGAACGCATCGAGAATTCAGTTTTCGACCGAACGTTTTCCGTGTGGCCATTTTTAGCGGAAACTATGTATAGAAATCCTCTCTATAAGCATGAGACTGAAAAGGTAGATGTAATTTTGGTATACACTGGCAGATTGAAGGGTAATGCTAATGATTTTTCTGATTTCTGTTTTTTAGGTTCTTTGGCCGGCGCACAGTGTGCGGTTTTTATATTTTTGGTCTGATGTATACCTTGGTAGTTTAGGCAACAAAAACGGAACTGATCTGCCATTACTAAGTAATGAAAGACAGAGCGGACACAATGGTGAATTAAGAAGTACCACTAATAGAAATGTACCACATACTCACATTTCATTGCTCCCACAACCAATAGGCCTCATGGCGAAGACACGATCTTCTGAAGACTTAACAGCGAACGAGTTG ..................................................................................................................................................................................................................................................................................................................................................((((((((((....(((.........(((((..........)))))...(((....)))........)))...))))))))))................................................................ ...............................................................................................................................................................................................................................................................................................................................................336................................................................................................435................................................ | Size | Perfect hit | Total Norm | Perfect Norm | V107 | V060 | V108 |
|---|---|---|---|---|---|---|---|
| ................................................................................................................TTTTTAGCGGAAACTATG................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ....................................................................................................................................................................................................................................................................................................................AACAAAAACGGAACTGATCTGCC.......................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................GAACGCATCGAGAATTCAGT................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ....................................................................................................................................................................................................................................................................................TTTGGTCTGATGTATACCTTGGTAGT....................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACACGATCTTCTGAAGACTTAACAGC.......... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................ACAGAGCGGACACAATGGT..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................GTAATGAAAGACAGAGCGGAC............................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ........................................................................................................................................................................................................................................................................GGTTTTTATATTTTTGGTCT......................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| GATCAAACTGGCGCAACATTCGTTGTCCTGTCTCTTCGGCACGTTCCTATGTAATTCGCTCAGAGAACGCATCGAGAATTCAGTTTTCGACCGAACGTTTTCCGTGTGGCCATTTTTAGCGGAAACTATGTATAGAAATCCTCTCTATAAGCATGAGACTGAAAAGGTAGATGTAATTTTGGTATACACTGGCAGATTGAAGGGTAATGCTAATGATTTTTCTGATTTCTGTTTTTTAGGTTCTTTGGCCGGCGCACAGTGTGCGGTTTTTATATTTTTGGTCTGATGTATACCTTGGTAGTTTAGGCAACAAAAACGGAACTGATCTGCCATTACTAAGTAATGAAAGACAGAGCGGACACAATGGTGAATTAAGAAGTACCACTAATAGAAATGTACCACATACTCACATTTCATTGCTCCCACAACCAATAGGCCTCATGGCGAAGACACGATCTTCTGAAGACTTAACAGCGAACGAGTTG ..................................................................................................................................................................................................................................................................................................................................................((((((((((....(((.........(((((..........)))))...(((....)))........)))...))))))))))................................................................ ...............................................................................................................................................................................................................................................................................................................................................336................................................................................................435................................................ | Size | Perfect hit | Total Norm | Perfect Norm | V107 | V060 | V108 |
|---|