|
(2) 2OME.ip |
(2) 2OME_MUTANT.ip |
(3) AGO1.ip |
(1) AGO2.ip |
(2) AGO2.mut |
(5) BODY |
(1) DCR2.mut |
(1) DISC |
(2) EMBRYO |
(12) HEAD |
(1) LOQS.mut |
(1) OSS |
(2) OTHER.ip |
(3) OTHER.mut |
(5) OVARY |
(1) PIWI.ip |
(2) R2D2.mut |
(2) S2 |
(1) S2_R |
(20) TOTAL_RNA.ip |
(3) WHOLE |
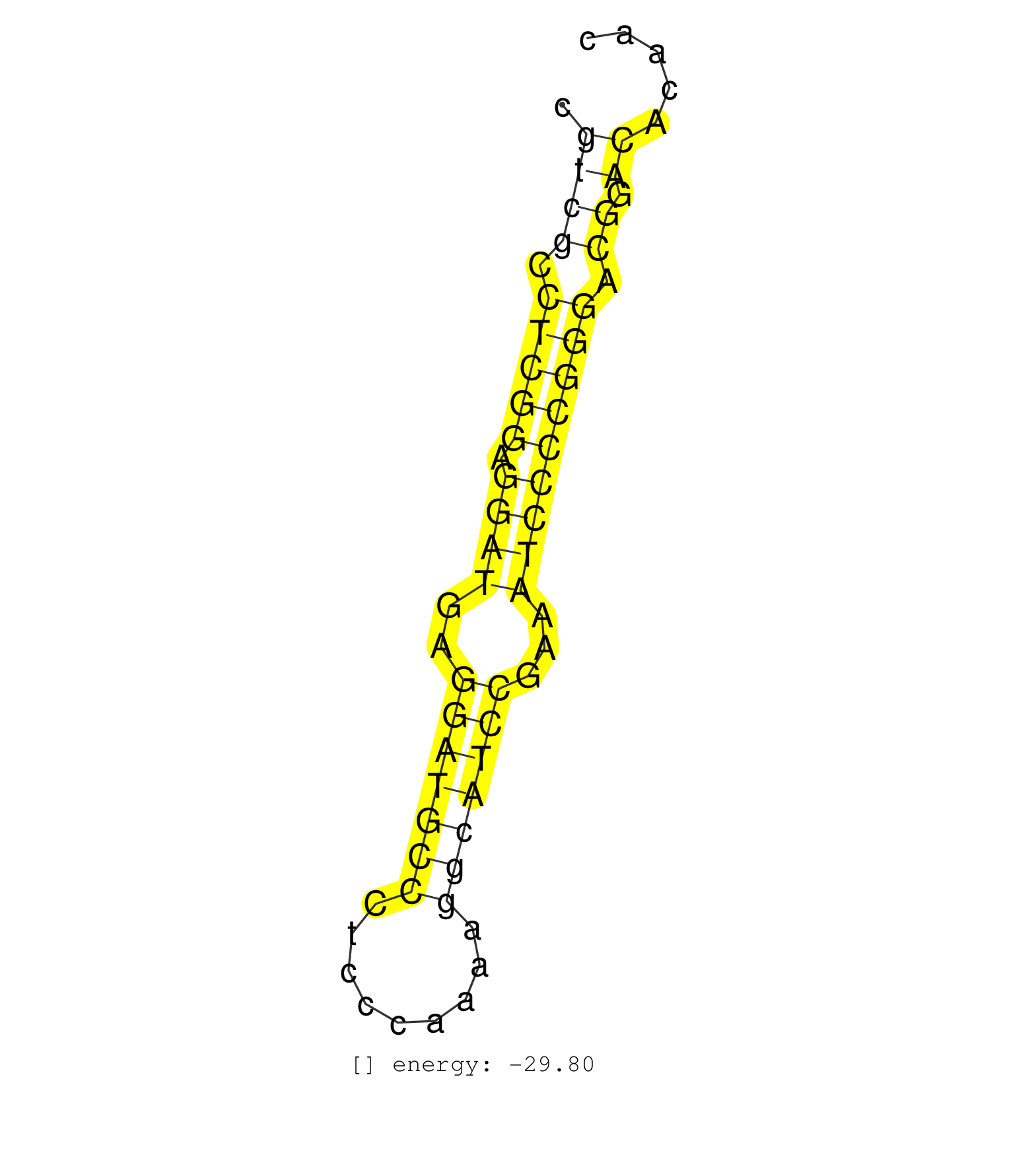
| GGGGGGGACTTTACCCCCCACACTCGAGAAACAGCAGAATCACGCAGGGCTTCCAGTGCGTCGCCTCGGAGGATGAGGATGCCCTCCCAAAAGGCATCCGAAATCCCCGGGACGGACACAACCCGTATTTAGGTGATTGGTCAAGGAATTGGACTGAGGAGACACAAGAAGA ...........................................................((((.(((((.((((..(((((((.........)))))))...))))))))).)).))....................................................... ..........................................................59.............................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | SRR031696:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V082:GSM609238 IP:total RNA embryo 14-24hr | SRR065155:GSM577962 IP:piwi ies.. | V142: IP:total RNA Oxidation_female_body | V086: IP:total RNA female body, aged | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V015: IP:total RNA DreRFHV148h | S5:GSM275691 IP:total RNA imaginal disc | SRR298536:GSM744629 IP:total RNA erozygote.. | V084:GSM609224 IP:total RNA female, one day | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | S23:GSM399106 IP:total RNA female body #2 | SRR010960:SRX002245 IP:2'Ome in mutant wt, oxidized | V066:GSM609233 IP:ago1 MUT:r2d2 r2d2[1], ovary, AGO1IP | SRR298711:GSM744619 SETDB1 Mutant.. | GSM467731: MUT:loqs Dmel_loq_sRNAseq | SRR031702:GSM466495 IP:2Ome flies.. | V091: IP:total RNA fGS/OSS total | SRR341116:GSM786605 IP:Luciferase MUT:Oxidized pe: Schneider 2 (S2) cellsoxidation.. | V081:GSM609229 IP:total RNA embryo 2-6hr | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR031697:GSM466490 IP:total RNA Total small RNAs from dcr-2 heterozygous flies | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR031698:GSM466491 IP:2Ome flies.. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................ATCCGAAATCCCCGGGACGGACA...................................................... | 23 | 1 | 24.00 | 24.00 | 19.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CCGAAATCCCCGGGACGGACA...................................................... | 21 | 1 | 10.00 | 10.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CATCCGAAATCCCCGGGACGGAC....................................................... | 23 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................ATCCGAAATCCCCGGGACGGA........................................................ | 21 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GGCATCCGAAATCCCCGGGACGGACA...................................................... | 26 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GGCATCCGAAATCCCCGGGACGGACAC..................................................... | 27 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GCATCCGAAATCCCCGGGACGGAC....................................................... | 24 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AATTGGACTGAGGAGAagcc...... | 20 | agcc | 3.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................ATCCGAAATCCCCGGGACGGAC....................................................... | 22 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AATTGGACTGAGGAGAagc....... | 19 | agc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CTGAGGAGACACAAGAAG. | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CATCCGAAATCCCCGGGACGGACAC..................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CGAAATCCCCGGGACGGACA...................................................... | 20 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GGCATCCGAAATCCCCGGGACGGAC....................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CATCCGAAATCCCCGGGACGGACA...................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAAGGCATCCGAAATCCCCGGGACGGACAC..................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CGTCGCCTCGGAGGATGAGG.............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AATTGGACTGAGGAGAat........ | 18 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................ACAACCCGTATTTAGGTGATT.................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GCCTCGGAGGATGAGaag............................................................................................ | 18 | aag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GCATCCGAAATCCCCGGGACGGACA...................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CGCCTCGGAGGATGAGGA............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GCGTCGCCTCGGAGGATGAGGAT............................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................GAGGATGAGGATGCCtac...................................................................................... | 18 | tac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................ATCCGAAATCCCCGGGACG.......................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CCGAAATCCCCGGGACGGAC....................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCTCGGAGGATGAGGATGCCC........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................ATCCGAAATCCCCGGGACGGtca...................................................... | 23 | tca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................AGAAACAGCAGAATCACGCAGGGC.......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TTTAGGTGATTGGTCAAGGAta....................... | 22 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TCCGAAATCCCCGGGACGGACA...................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGGAATTGGACTGAGGAGACAC....... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CGAAATCCCCGGGACGGAC....................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CGAGAAACAGCAGAATattg................................................................................................................................ | 20 | attg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................GGCTTCCAGTGCGTCGCCTCGGAGG.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ATTGGACTGAGGAGACACAAG.... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GAAATCCCCGGGACGGACA...................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AGTGCGTCGCCTCGGAGGATGAGG.............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............ACCCCCCACACTC................................................................................................................................................... | 13 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - |
| ..............................................................GCCTCGGAGGAT.................................................................................................. | 12 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| ...................................................................................................GAAATCCCCGGGA............................................................ | 13 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TAGGTGATTGGT............................... | 12 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| ..................................................................................................................................................AATTGGACTGAGGA............ | 14 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - |
| ..........................AGAAACAGCAGAAT.................................................................................................................................... | 14 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - |
| ..........................................................................................................CCGGGACGGACA...................................................... | 12 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - |
| ...................................................................................................................................................ATTGGACTGAGGA............ | 13 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - |
| ....................................................................................................................CACAACCCGTA............................................. | 11 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| GGGGGGGACTTTACCCCCCACACTCGAGAAACAGCAGAATCACGCAGGGCTTCCAGTGCGTCGCCTCGGAGGATGAGGATGCCCTCCCAAAAGGCATCCGAAATCCCCGGGACGGACACAACCCGTATTTAGGTGATTGGTCAAGGAATTGGACTGAGGAGACACAAGAAGA ...........................................................((((.(((((.((((..(((((((.........)))))))...))))))))).)).))....................................................... ..........................................................59.............................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | SRR031696:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V082:GSM609238 IP:total RNA embryo 14-24hr | SRR065155:GSM577962 IP:piwi ies.. | V142: IP:total RNA Oxidation_female_body | V086: IP:total RNA female body, aged | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V015: IP:total RNA DreRFHV148h | S5:GSM275691 IP:total RNA imaginal disc | SRR298536:GSM744629 IP:total RNA erozygote.. | V084:GSM609224 IP:total RNA female, one day | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | S23:GSM399106 IP:total RNA female body #2 | SRR010960:SRX002245 IP:2'Ome in mutant wt, oxidized | V066:GSM609233 IP:ago1 MUT:r2d2 r2d2[1], ovary, AGO1IP | SRR298711:GSM744619 SETDB1 Mutant.. | GSM467731: MUT:loqs Dmel_loq_sRNAseq | SRR031702:GSM466495 IP:2Ome flies.. | V091: IP:total RNA fGS/OSS total | SRR341116:GSM786605 IP:Luciferase MUT:Oxidized pe: Schneider 2 (S2) cellsoxidation.. | V081:GSM609229 IP:total RNA embryo 2-6hr | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR031697:GSM466490 IP:total RNA Total small RNAs from dcr-2 heterozygous flies | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR031698:GSM466491 IP:2Ome flies.. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TCGGAGGATGAGGATGCC......................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...tcaGACTTTACCCCCCACA...................................................................................................................................................... | 19 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................gcaAATTGGACTGAGGAGAC......... | 20 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....agcACTTTACCCCCCACA...................................................................................................................................................... | 18 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGGACGGACACAAC.................................................. | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ttaTTACCCCCCACACTC................................................................................................................................................... | 18 | tta | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .cggtGGACTTTACCCCCCAC....................................................................................................................................................... | 20 | cggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................cCCCCGGGACGGACACAACCCGTA............................................. | 24 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...tcgaACTTTACCCCCCACA...................................................................................................................................................... | 19 | tcga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TTTACCCCCCAC....................................................................................................................................................... | 12 | 13 | 0.38 | 0.38 | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.15 | 0.15 | - | - | - | - | - |
| ........................................................................................................CCCCGGGACGG......................................................... | 11 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................ATGCCCTCCCAA.................................................................................. | 12 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CTTTACCCCCCA........................................................................................................................................................ | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - |
| .............................................................CGCCTCGGAGGA................................................................................................... | 12 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - |
| ..........TTACCCCCCACA...................................................................................................................................................... | 12 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - |
| .............................................................................................................................................................GGAGACACAAGA... | 12 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - |
| .................................................................................CCCTCCCAAAAG............................................................................... | 12 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - |