|
(1) 2OME.ip |
(2) 2OME_MUTANT.ip |
(2) AGO1.ip |
(2) AGO2.mut |
(3) BODY |
(2) DCR2.mut |
(4) EMBRYO |
(14) HEAD |
(1) LOQS.mut |
(8) OVARY |
(1) PIWI.ip |
(3) R2D2.mut |
(2) S1 |
(1) S3 |
(24) TOTAL_RNA.ip |
(1) WHOLE |
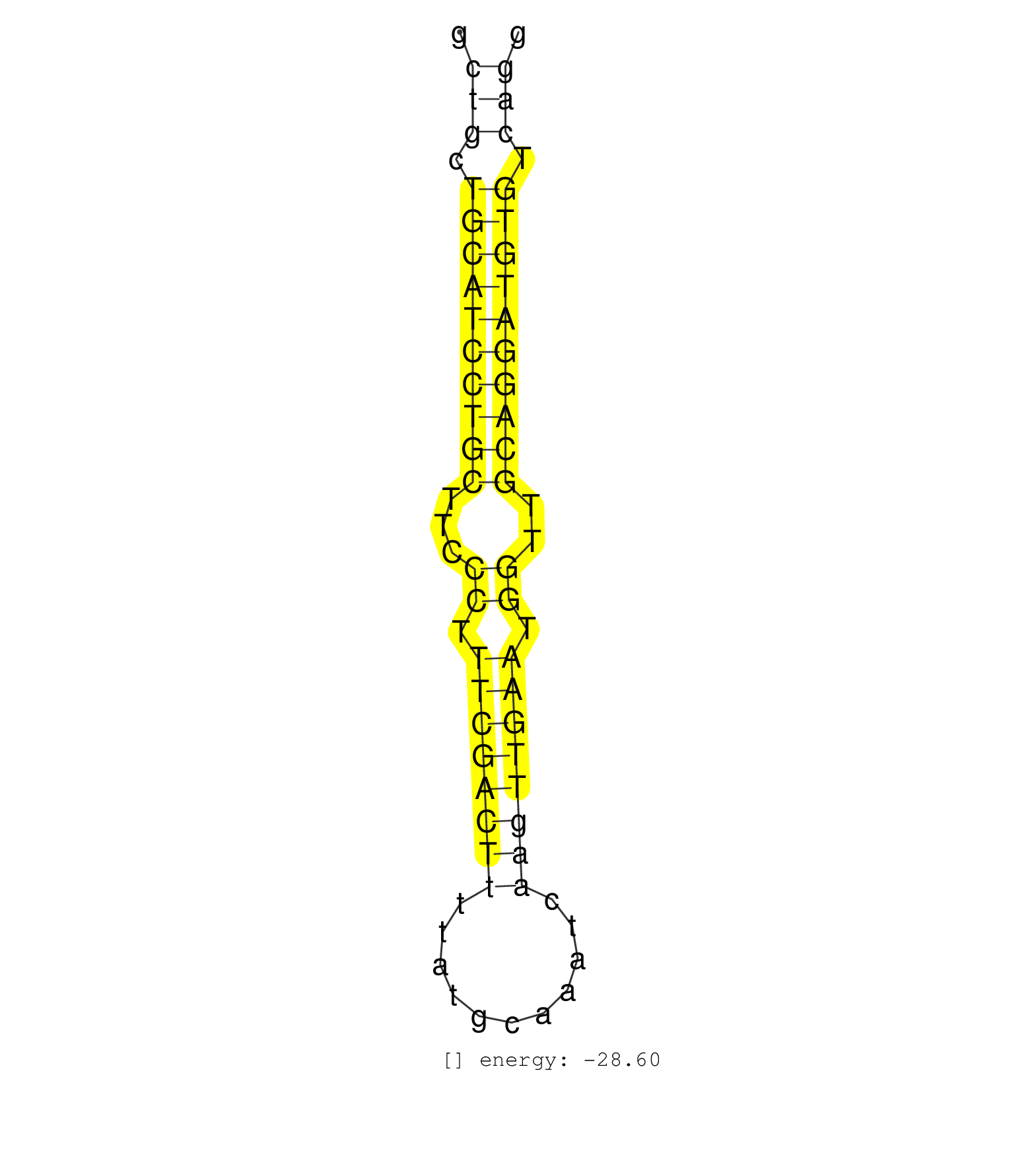
| TAAACTTAAATTCCCCCGAAGCCTCGAGATGTCGAGAAGTTCCGATTCCTGCTGCTAGGCTGCTGCATCCTGCTTCCCTTTCGACTTTTATGCAAATCAAGTTGAATGGTTGCAGGATGTGTCAGGTAGCTGTAGCTACCTCTAGACCTGCCTCGTAGGCATTTCTGTGCAAATTCCATATTTACTGCCA ...........................................................(((.((((((((((...((.((((((((...........)))))))).))..)))))))))).)))................................................................. ..........................................................59.................................................................126.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR031697:GSM466490 IP:total RNA Total small RNAs from dcr-2 heterozygous flies | S9:GSM286604 IP:total RNA 0-1h #3 (7) | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | V082:GSM609238 IP:total RNA embryo 14-24hr | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR317117:GSM767601 IP:total RNA ld flies.. | GSM467731: MUT:loqs Dmel_loq_sRNAseq | V146: IP:total RNA S1 cell | S6: IP:total RNA 0-1,2-6,6-10h embryo | SRR031699:GSM466492 IP:total RNA Total small RNAs from dcr-2 homozygous flies | SRR031698:GSM466491 IP:2Ome flies.. | S3:GSM286602 IP:total RNA male body | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | V032: IP:total RNA S1 cell | V013:GSM609251 IP:total RNA aged female head | SRR317116:GSM767600 IP:total RNA 5-6 days old flies.. | SRR031700:GSM466493 IP:2'Ome in mutant MUT:dcr2 lies.. | SRR351332:GSM811191 IP:total RNA Nibbler ygenotype: control wild type (w, 5905).. | SRR065151:GSM577958 IP:piwi Piwi bound piRNAs from tj GAL4 > RNAi-aub ovaries | V074: IP:total RNA S3 | V0632: IP:ago1 wt ovary AGO1-IP, reseq | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V097:GSE24545 IP:total RNA CS ovary total RNA | S22:GSM322533 IP:total RNA female head #1 | V077: IP:total RNA cold, female head | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR298711:GSM744619 SETDB1 Mutant.. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................TTTTATGCAAATCAAtata...................................................................................... | 19 | tata | 46.00 | 0.00 | 15.00 | 8.00 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................TTGAATGGTTGCAGGATGTGT.................................................................... | 21 | 1 | 9.00 | 9.00 | - | - | - | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................TTGAATGGTTGCAGGATGTGTC................................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGCATCCTGCTTCCCTTTCGACT........................................................................................................ | 23 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................GGTTGCAGGATGTGT.................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................TTTCTGTGCAAATTCCATATT........ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TTGCAGGATGTGTCActca.............................................................. | 19 | ctca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAtgta...................................................................................... | 19 | tgta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGCATCCTGCTTCCCTTTCGACTTTTAT................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAtcta...................................................................................... | 19 | tcta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATGGTTGCAGGATGTa..................................................................... | 16 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAcata...................................................................................... | 19 | cata | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGCATCCTGCTTCCCTTTCGAC......................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................AGAAGTTCCGATTCCTGCTGC....................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CCGAAGCCTCGAGATGTCGA........................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAttta...................................................................................... | 19 | ttta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CCCGAAGCCTCGAGATGTCG............................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TTGCAGGATGTGTCAccc............................................................... | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCCATATTTACTGC.. | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGTTGCAGGATGTGTCAGGTA.............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAtatc...................................................................................... | 19 | tatc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTATGCAAATCAAaat....................................................................................... | 18 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TTCCGATTCCTGC.......................................................................................................................................... | 13 | 7 | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| ......................................GTTCCGATTCCTG........................................................................................................................................... | 13 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ...................AGCCTCGAGATG............................................................................................................................................................... | 12 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........ATTCCCCCGAAG......................................................................................................................................................................... | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CCGAAGCCTCGA................................................................................................................................................................... | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGTTGCAGGAT........................................................................ | 12 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - |
| ..........................................................GCTGCTGCATCCT....................................................................................................................... | 13 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - |
| ....CTTAAATTCCCC.............................................................................................................................................................................. | 12 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - |
| .......................................................................................................................................CTACCTCTAGA............................................ | 11 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TAAACTTAAATTCCCCCGAAGCCTCGAGATGTCGAGAAGTTCCGATTCCTGCTGCTAGGCTGCTGCATCCTGCTTCCCTTTCGACTTTTATGCAAATCAAGTTGAATGGTTGCAGGATGTGTCAGGTAGCTGTAGCTACCTCTAGACCTGCCTCGTAGGCATTTCTGTGCAAATTCCATATTTACTGCCA ...........................................................(((.((((((((((...((.((((((((...........)))))))).))..)))))))))).)))................................................................. ..........................................................59.................................................................126.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR031697:GSM466490 IP:total RNA Total small RNAs from dcr-2 heterozygous flies | S9:GSM286604 IP:total RNA 0-1h #3 (7) | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | V082:GSM609238 IP:total RNA embryo 14-24hr | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR317117:GSM767601 IP:total RNA ld flies.. | GSM467731: MUT:loqs Dmel_loq_sRNAseq | V146: IP:total RNA S1 cell | S6: IP:total RNA 0-1,2-6,6-10h embryo | SRR031699:GSM466492 IP:total RNA Total small RNAs from dcr-2 homozygous flies | SRR031698:GSM466491 IP:2Ome flies.. | S3:GSM286602 IP:total RNA male body | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | V032: IP:total RNA S1 cell | V013:GSM609251 IP:total RNA aged female head | SRR317116:GSM767600 IP:total RNA 5-6 days old flies.. | SRR031700:GSM466493 IP:2'Ome in mutant MUT:dcr2 lies.. | SRR351332:GSM811191 IP:total RNA Nibbler ygenotype: control wild type (w, 5905).. | SRR065151:GSM577958 IP:piwi Piwi bound piRNAs from tj GAL4 > RNAi-aub ovaries | V074: IP:total RNA S3 | V0632: IP:ago1 wt ovary AGO1-IP, reseq | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V097:GSE24545 IP:total RNA CS ovary total RNA | S22:GSM322533 IP:total RNA female head #1 | V077: IP:total RNA cold, female head | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR298711:GSM744619 SETDB1 Mutant.. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................aagTGCTTCCCTTTCGAC......................................................................................................... | 18 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................GTCGAGAAGTTCCGATTC.............................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................GCCTCGAGATGTCGAGAAGTTCCGA................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................cgggCTGCTTCCCTTTCGACT........................................................................................................ | 21 | cggg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................GATGTCGAGAAGTT..................................................................................................................................................... | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................gacCTTCCCTTTCGACTTTTA.................................................................................................... | 21 | gac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................AGATGTCGAGAAGTTCCGATTC.............................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................tCTGCATCCTGCTTCC................................................................................................................. | 16 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CCTGCTGCTAGG................................................................................................................................... | 12 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TACCTCTAGAC........................................... | 11 | 12 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - |
| .......................................................................................................................................CTACCTCTAGA............................................ | 11 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |