|
(2) AGO1.ip |
(2) AGO2.ip |
(2) AGO2.mut |
(5) BODY |
(2) DCR2.mut |
(1) G2 |
(6) HEAD |
(2) LOQS.mut |
(1) OSC |
(2) OTHER.ip |
(7) OTHER.mut |
(6) OVARY |
(1) PUPAE |
(4) R2D2.mut |
(1) S1 |
(6) S2 |
(5) S2_R |
(1) S3 |
(32) TOTAL_RNA.ip |
(4) WHOLE |
(1) WING |
(2) mbn2 |
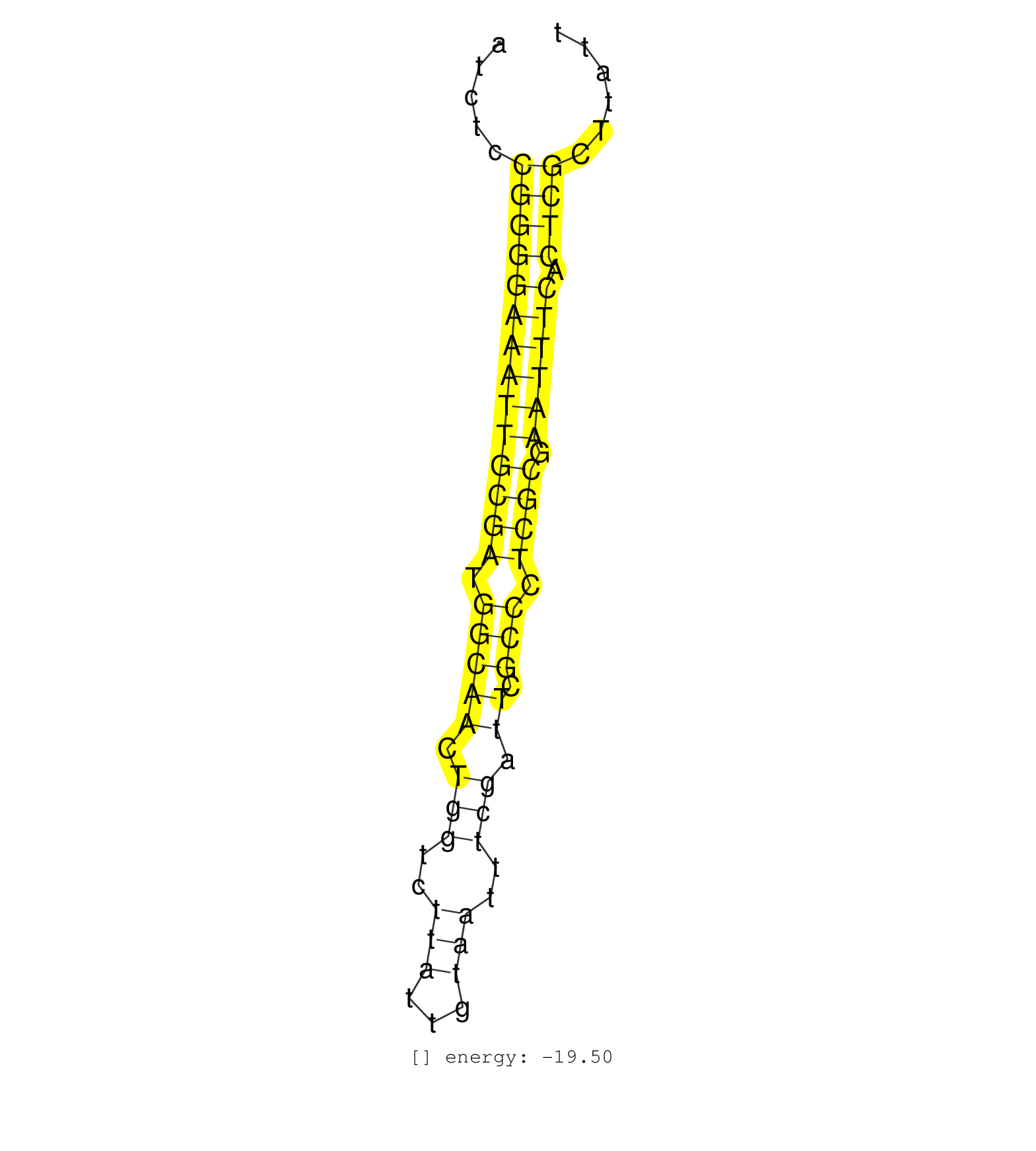
| CAATTCGTCAACCTCTAACCCAATCCTTATCGCCGCGAATATGTCCAACCTACAGCAGAAATCTCCGGGGAAATTGCGATGGCAACTGGTCTTATTGTAATTTCGATTCGCCCTCGCGAATTTCACTCGCTTATTTCTGACCCAATTTCCCGAATGCATCACTTCCTCCATATCTTCCCGCGGAGGAGAAT .................................................................((((((((((((((.(((((.(((..(((...)))..))).)).))).)))).)))))).)))).............................................................. ............................................................61........................................................................135...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | S32:GSM609242 IP:total RNA s2+48 #2 | S31:GSM609241 IP:total RNA s2+48 #1 | M067: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | V146: IP:total RNA S1 cell | V148: IP:total RNA mbn2 | SRR029031:GSM430033 MUT:loqs loqs-ORF knockdown | M064: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | SRR341115:GSM786604 IP:Nibbler MUT:Unoxidized Schneider 2 (S2) cellsoxidation.. | V127: IP:total RNA G2 | SRR317115:GSM767599 IP:total RNA old flies.. | V085: IP:total RNA CME W2 wing disc | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V0892:GSM628272 IP:total RNA MUT:ago2 ago2[414] ovary total RNA | SRR065801:GSM593298 IP:total RNA zuc_het(H-Y)_ovaries | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V144: IP:total RNA OSC | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | M063: IP:AGO1 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V128: IP:total RNA S3 | SRR317117:GSM767601 IP:total RNA ld flies.. | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | V145: IP:total RNA S2-DRSC | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V073: IP:total RNA mbn2 | V141: IP:total RNA Heat_female_body | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | S28:GSM360262 IP:total RNA 0-2d pupae | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | V086: IP:total RNA female body, aged | V138: IP:total RNA Male cold body | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................CGGGGAAATTGCGATGGCAACT........................................................................................................ | 22 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................CGGGGAAATTGCGATGGC............................................................................................................ | 18 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCGCCCTCGCGAATTTCACTCGCT............................................................ | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................CTCCATATCTTCCCG........... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCGCCCTCGCGAATTTCACTCG.............................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CGGGGAAATTGCGATGGCAACTGGTC.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGCATCACTTCCTCCATATCT................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TGTCCAACCTACAGCAGAAATCTC.............................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .AATTCGTCAACCTCTAACCCA......................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CATCACTTCCTCCATcc.................. | 17 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................AATATGTCCAACCTACAGCAG..................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CGGGGAAATTGCGATGGCAAtt........................................................................................................ | 22 | tt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAATTTCGATTCGCCCTCGCGt........................................................................ | 22 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CGCTTATTTCTGACCCAATT............................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCGCCCTCGCGAATTTCACTCGC............................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TGTCCAACCTACAGCAGAA................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GCAGAAATCTCCGGGGAAATT.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATCTCCGGGGAAATTGCGATGGC............................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAATTTCGATTCGCCCTCGCGAATT..................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCGCCCTCGCGAATTTCACTCGCc............................................................ | 24 | c | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CGTCAACCTCTAACCCAATCC..................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TCAACCTCTAACCCAATCCT.................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................ATTGCGATGGCAACTGGatgg.................................................................................................. | 21 | atgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................GGTCTTATTGTA............................................................................................ | 12 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................ATCTTCCCGCG......... | 11 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - |
| .......................................................................................................................ATTTCACTCGCTT........................................................... | 13 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 |
| CAATTCGTCAACCTCTAACCCAATCCTTATCGCCGCGAATATGTCCAACCTACAGCAGAAATCTCCGGGGAAATTGCGATGGCAACTGGTCTTATTGTAATTTCGATTCGCCCTCGCGAATTTCACTCGCTTATTTCTGACCCAATTTCCCGAATGCATCACTTCCTCCATATCTTCCCGCGGAGGAGAAT .................................................................((((((((((((((.(((((.(((..(((...)))..))).)).))).)))).)))))).)))).............................................................. ............................................................61........................................................................135...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | S32:GSM609242 IP:total RNA s2+48 #2 | S31:GSM609241 IP:total RNA s2+48 #1 | M067: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | V146: IP:total RNA S1 cell | V148: IP:total RNA mbn2 | SRR029031:GSM430033 MUT:loqs loqs-ORF knockdown | M064: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | SRR341115:GSM786604 IP:Nibbler MUT:Unoxidized Schneider 2 (S2) cellsoxidation.. | V127: IP:total RNA G2 | SRR317115:GSM767599 IP:total RNA old flies.. | V085: IP:total RNA CME W2 wing disc | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V0892:GSM628272 IP:total RNA MUT:ago2 ago2[414] ovary total RNA | SRR065801:GSM593298 IP:total RNA zuc_het(H-Y)_ovaries | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V144: IP:total RNA OSC | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | M063: IP:AGO1 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V128: IP:total RNA S3 | SRR317117:GSM767601 IP:total RNA ld flies.. | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | V145: IP:total RNA S2-DRSC | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V073: IP:total RNA mbn2 | V141: IP:total RNA Heat_female_body | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | S28:GSM360262 IP:total RNA 0-2d pupae | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | V086: IP:total RNA female body, aged | V138: IP:total RNA Male cold body | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................aagGAATTTCACTCGCTTA.......................................................... | 19 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................CTTCCTCCATATCTTCCCG........... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................tTCCATATCTTCCCGCGGAGGA.... | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................gcaCATCACTTCCTCCAT.................... | 18 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................aaACCCAATTTCCCGAAT.................................... | 18 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ACTTCCTCCATATCTTCC............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AATTGCGATGGCAACTGGTCTTAT................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GACCCAATTTCCCGAATGCAT................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................cctCATCACTTCCTCCAT.................... | 18 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CAACCTCTAACCCAATCCTTA.................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................CCATATCTTCCCGCGGAGGAG... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................ACTCGCTTATTTCTGACCCAA.............................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................TTCGCCCTCGCGAATTTCACTCGCTTA.......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................aCCGAATGCATCACTTCCTCCA..................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................cgggTATTTCTGACCCAATTT........................................... | 21 | cggg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GTCCAACCTACAGCAGAAATC................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................aaaAATGCATCACTTCCT........................ | 18 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATTCGCCCTCGCG......................................................................... | 13 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAACCCAATCCTTA.................................................................................................................................................................. | 14 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........CCTCTAACCCAAT....................................................................................................................................................................... | 13 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........AACCTCTAACCC.......................................................................................................................................................................... | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - |
| .....................................................................................................................................................CCGAATGCATCA.............................. | 12 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - |
| ...................................................................................................................................................................TCCTCCATATCTT............... | 13 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - |