|
(2) 1182-4H |
(2) 2OME.ip |
(2) 2OME_MUTANT.ip |
(1) AGO1.ip |
(1) AGO1.mut |
(1) AGO2.ip |
(1) AGO2.mut |
(4) BODY |
(1) CMEW1 Cl.8+ cell |
(1) DCR2.mut |
(4) EMBRYO |
(8) HEAD |
(1) KC |
(1) LARVAE |
(2) LOQS.mut |
(1) ML_CELL_LINE |
(1) OSC |
(2) OSS |
(1) OTHER.ip |
(7) OTHER.mut |
(6) OVARY |
(1) PIWI.ip |
(1) R2D2.mut |
(6) S2 |
(3) S2_R |
(30) TOTAL_RNA.ip |
(1) WHOLE |
(2) mbn2 |
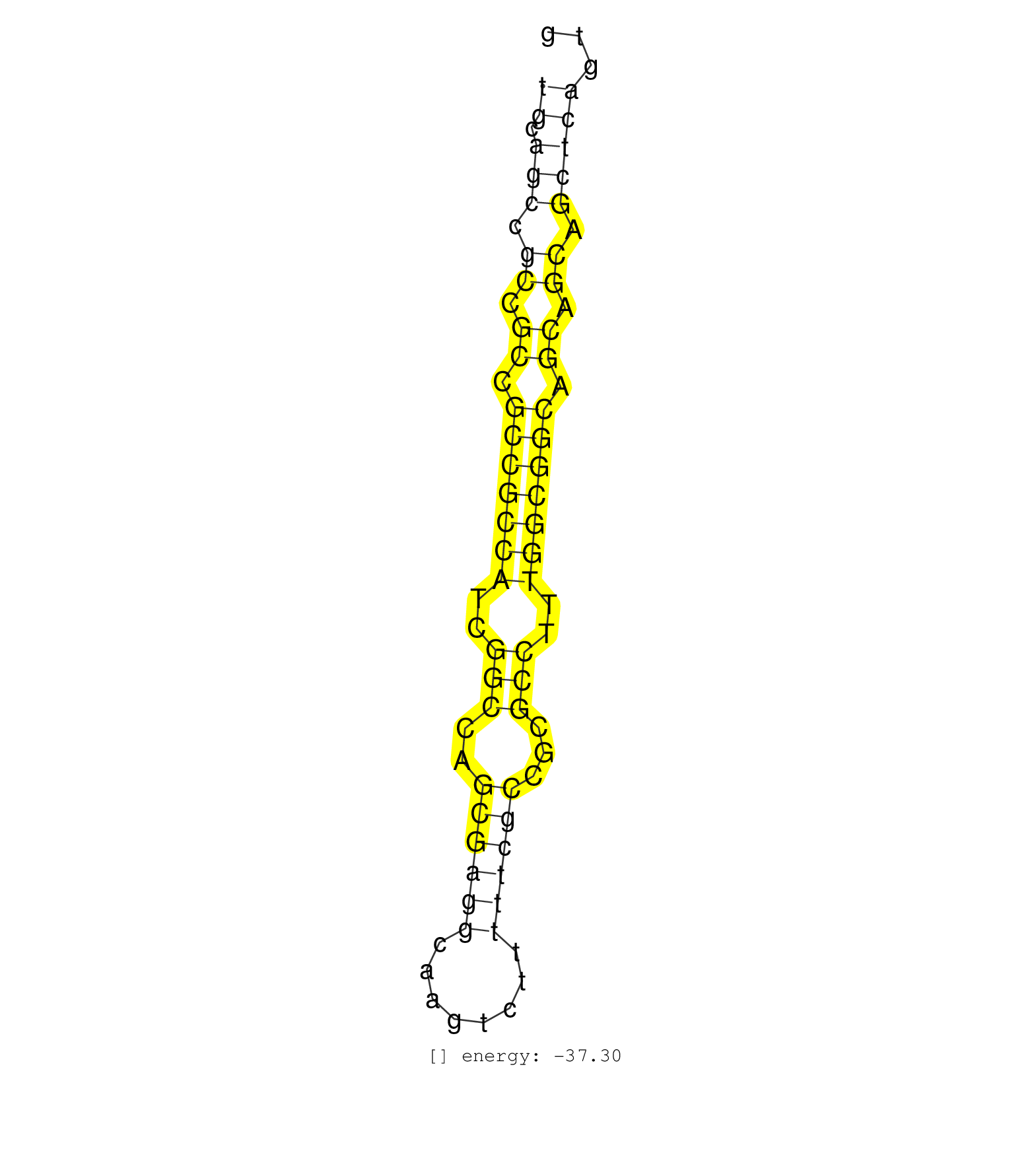
| ATGTCCCGAGTGCGAAAAGTTTCGCTGCATGTGTGAGCGCCCGGCAAGTGTGCAGCCGCCGCCGCCGCCATCGGCCAGCGAGGCAAGTCTTTTTCGCCGCGCCTTTGGCGGCAGCAGCAGCTCAGTGGCCACCAGTAGCGCCCAGTCCACGCCCACCAGCGATTGCAGCTCCGCCGG ..................................................((.(((.((.((.(((((((..(((..((((((........))))))...)))..))))))).)).)).)))))..................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR065806:GSM593303 IP:piwi MUT:squ Piwi-IP_squ_mut_ovaries | V079: IP:2'Ome in mutant Oxidation, female head | V147: IP:total RNA 1182-4H cell | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | GSM379052: IP:total RNA Aub Heterozygote | S33:GSM399110 IP:total RNA KC-48 #2 | M064: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR032093:GSM442733 MUT:ago1 ago1 knockdown | S32:GSM609242 IP:total RNA s2+48 #2 | SRR298537:GSM744630 MUT:Vret ret Mutant.. | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V137: IP:total RNA Male aged head | V037: IP:total RNA Felix sample +mirtrons | V026:GSM609221 IP:total RNA 1182-4H cell | GSM343287: IP:total RNA Drosophila Toll 10b mutant embryos | V001:GSM609239 IP:total RNA MUT:IR IR- | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V144: IP:total RNA OSC | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | OSS8:GSM385822 IP:total RNA OSS_s8 | V098:GSE24544 IP:total RNA MUT:dcr2 dcr-2[L811fsX] ovary total RNA | V148: IP:total RNA mbn2 | SRR029031:GSM430033 MUT:loqs loqs-ORF knockdown | S7:GSM364902 IP:total RNA 12-24hr embryo | V130: IP:total RNA ML-DmBG3-c2 | V140: IP:total RNA Dessication_female_body | SRR014273:GSM327622 IP:total RNA Ovary_rep1_Har_P | V073: IP:total RNA mbn2 | SRR031702:GSM466495 IP:2Ome flies.. | SRR031698:GSM466491 IP:2Ome flies.. | S3:GSM286602 IP:total RNA male body | S25:GSM360256 IP:total RNA 1st instar #1 | S15:GSM272652 IP:total RNA S2 -48 Biological Replicate #1 | S22:GSM322533 IP:total RNA female head #1 | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | OSS6:GSM385748 IP:total RNA OSS_s6 | V081:GSM609229 IP:total RNA embryo 2-6hr | M066: IP:AGO1 MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR298711:GSM744619 SETDB1 Mutant.. | SRR031699:GSM466492 IP:total RNA Total small RNAs from dcr-2 homozygous flies | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................CCGCCGCCGCCATCGGCCAGCG................................................................................................. | 22 | 1 | 12.00 | 12.00 | - | 1.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ..........................................................CCGCCGCCGCCATCGGCCAGC.................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GTGAGCGCCCGGCAAGTGTGCAGC......................................................................................................................... | 24 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGCCGCCGCCATCGGCCAGCG................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCGCGCCTTTGGCGGCAGCAGCA.......................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGCCGCCGCCATCGGCCAGC.................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................GCAGCCGCCGCCGCCGCCgcaa........................................................................................................ | 22 | gcaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCGCGCCTTTGGCGGCAGCAGCAG......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CGCCTTTGGCGGCAGCggg........................................................... | 19 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTTGGCGGCAGCAGCgcaa....................................................... | 19 | gcaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GGCGGCAGCAGCAGCTCAGTG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................TGCATGTGTGAGCGCCCGGC.................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CCGCCGCCGCCATCGGCCAGCGt................................................................................................ | 23 | t | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CCGCCGCCGCCATCGGCCAGa.................................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCTTTGGCGGCAGCAGtag......................................................... | 19 | tag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................TGCATGTGTGAGCGCCCGGCAAGTGT.............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CAGCAGCTCAGTGGCaga............................................. | 18 | aga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGCCGCCGCCATCGGCCAGCGt................................................................................................ | 22 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTTTGGCGGCAGCAGCAGCTCt..................................................... | 22 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCGCCATCGGCCAGCG................................................................................................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GCCGCCATCGGCCAGCGAGGa............................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GCAGCAGCTCAGTGGtag.............................................. | 18 | tag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCCACGCCCACCAGCGATTGCAGC........ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCTTTGGCGGCAGC.............................................................. | 14 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| ......................................GCCCGGCAAGTG............................................................................................................................... | 12 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - |
| ...................................................................................CAAGTCTTTTTC.................................................................................. | 12 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
| ATGTCCCGAGTGCGAAAAGTTTCGCTGCATGTGTGAGCGCCCGGCAAGTGTGCAGCCGCCGCCGCCGCCATCGGCCAGCGAGGCAAGTCTTTTTCGCCGCGCCTTTGGCGGCAGCAGCAGCTCAGTGGCCACCAGTAGCGCCCAGTCCACGCCCACCAGCGATTGCAGCTCCGCCGG ..................................................((.(((.((.((.(((((((..(((..((((((........))))))...)))..))))))).)).)).)))))..................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR065806:GSM593303 IP:piwi MUT:squ Piwi-IP_squ_mut_ovaries | V079: IP:2'Ome in mutant Oxidation, female head | V147: IP:total RNA 1182-4H cell | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | GSM379052: IP:total RNA Aub Heterozygote | S33:GSM399110 IP:total RNA KC-48 #2 | M064: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | SRR032093:GSM442733 MUT:ago1 ago1 knockdown | S32:GSM609242 IP:total RNA s2+48 #2 | SRR298537:GSM744630 MUT:Vret ret Mutant.. | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V137: IP:total RNA Male aged head | V037: IP:total RNA Felix sample +mirtrons | V026:GSM609221 IP:total RNA 1182-4H cell | GSM343287: IP:total RNA Drosophila Toll 10b mutant embryos | V001:GSM609239 IP:total RNA MUT:IR IR- | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V144: IP:total RNA OSC | SRR341117:GSM786606 IP:Nibbler MUT:Oxidized Schneider 2 (S2) cellsoxidation.. | OSS8:GSM385822 IP:total RNA OSS_s8 | V098:GSE24544 IP:total RNA MUT:dcr2 dcr-2[L811fsX] ovary total RNA | V148: IP:total RNA mbn2 | SRR029031:GSM430033 MUT:loqs loqs-ORF knockdown | S7:GSM364902 IP:total RNA 12-24hr embryo | V130: IP:total RNA ML-DmBG3-c2 | V140: IP:total RNA Dessication_female_body | SRR014273:GSM327622 IP:total RNA Ovary_rep1_Har_P | V073: IP:total RNA mbn2 | SRR031702:GSM466495 IP:2Ome flies.. | SRR031698:GSM466491 IP:2Ome flies.. | S3:GSM286602 IP:total RNA male body | S25:GSM360256 IP:total RNA 1st instar #1 | S15:GSM272652 IP:total RNA S2 -48 Biological Replicate #1 | S22:GSM322533 IP:total RNA female head #1 | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | OSS6:GSM385748 IP:total RNA OSS_s6 | V081:GSM609229 IP:total RNA embryo 2-6hr | M066: IP:AGO1 MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR298711:GSM744619 SETDB1 Mutant.. | SRR031699:GSM466492 IP:total RNA Total small RNAs from dcr-2 homozygous flies | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................ccacCCGCCGCCGCCGCCA........................................................................................................... | 19 | ccac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................tctCGCCCACCAGCGATT............. | 18 | tct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................tcggTGGCGGCAGCAGCAGC........................................................ | 20 | tcgg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ccagAGCGCCCAGTCCACG.......................... | 19 | ccag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................gcTCCACGCCCACCAGCGA............... | 19 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................gggGGCGGCAGCAGCAGCTCA..................................................... | 21 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................gtCACCAGCGATTGCAG......... | 17 | gt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................gtccGCCGCCGCCGCCGCC............................................................................................................ | 19 | gtcc | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .................................................GTGCAGCCGCCGCCG................................................................................................................. | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |