|
(1) 1182-4H |
(3) 2OME_MUTANT.ip |
(3) AGO1.ip |
(1) AGO2.ip |
(3) AGO2.mut |
(10) BODY |
(1) CME L1 |
(1) CMEW1 Cl.8+ cell |
(4) DCR2.mut |
(1) EMBRYO |
(20) HEAD |
(2) LOQS.mut |
(3) ML_CELL_LINE |
(2) OTHER |
(4) OTHER.mut |
(11) OVARY |
(4) R2D2.mut |
(1) S2 |
(1) S2_NP |
(48) TOTAL_RNA.ip |
(9) WHOLE |
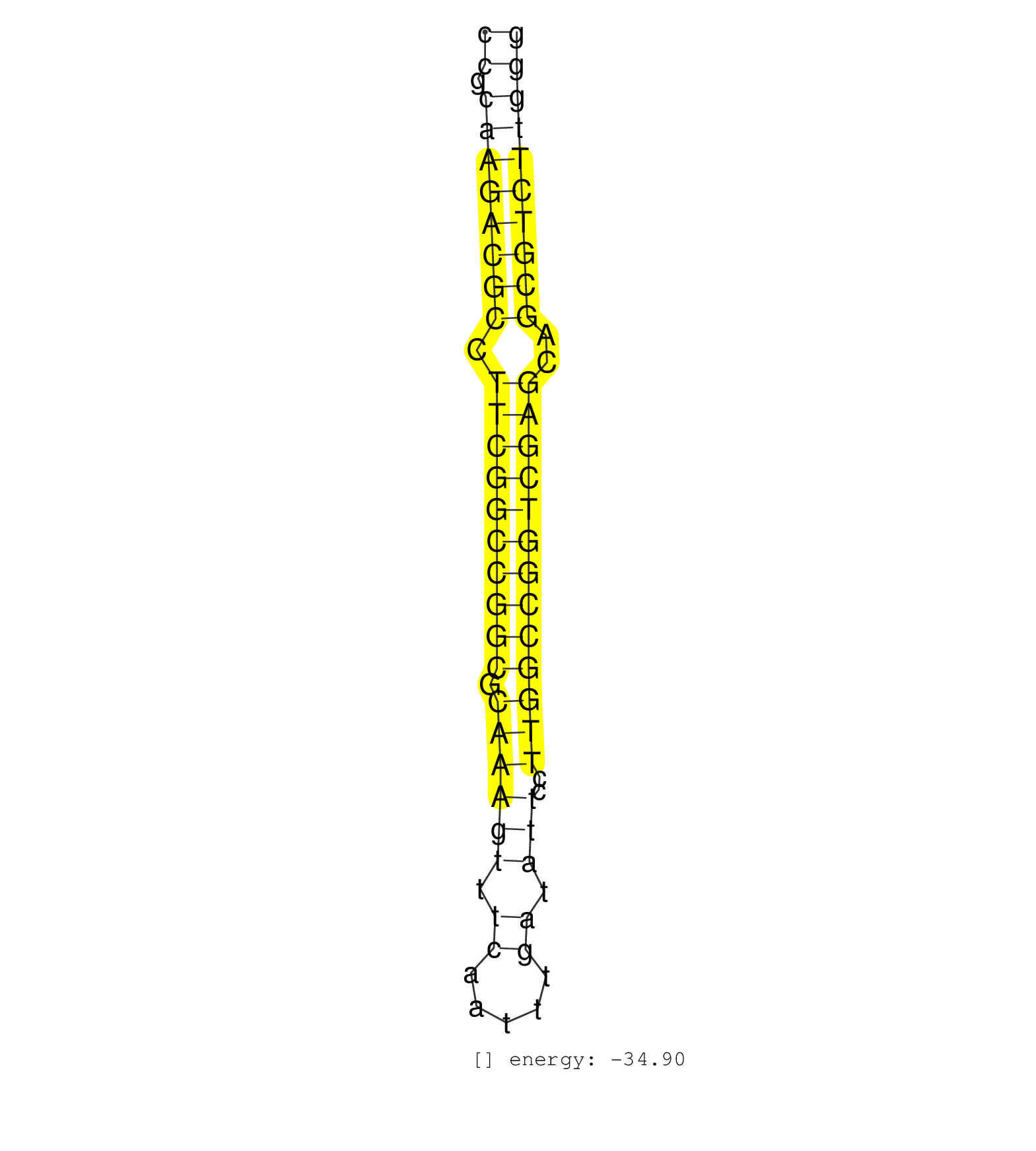
| TCGTCGAAAGGGTAATTGAAGAAGCGAAAACCTGGGTTATCCCTGCCACATGGCCACGTGCCCGCAAGACGCCTTCGGCCGGCGCAAAGTTTCAATTTGATATTCCTTGGCCGGTCGAGCAGCGTCTTGGGGTTGTTCGTGGATTAAGTTCGATTCCTATTCTGTGGCACTGTGGCTTTTCTCGCCGACAGGCTG .............................................................((.((((((((.((((((((((.((((((.((.....)).)))..)))))))))))))..))))))))))................................................................ .............................................................62...................................................................131.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | V137: IP:total RNA Male aged head | V138: IP:total RNA Male cold body | V136: IP:total RNA Male aged body | V141: IP:total RNA Heat_female_body | V142: IP:total RNA Oxidation_female_body | V139: IP:total RNA Cold_female_body | V077: IP:total RNA cold, female head | V080: IP:total RNA Starvation, female head | V140: IP:total RNA Dessication_female_body | V078: IP:total RNA Desiccation, female head | V090:GSM609234 IP:total RNA CS male total RNA | V079: IP:2'Ome in mutant Oxidation, female head | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V086: IP:total RNA female body, aged | S24:GSM399107 IP:total RNA male body #2 | V013:GSM609251 IP:total RNA aged female head | SRR317111:GSM767595 IP:total RNA : from 5-6 days old flies.. | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V098:GSE24544 IP:total RNA MUT:dcr2 dcr-2[L811fsX] ovary total RNA | V132: IP:total RNA ML-DmD32 | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR298712:GSM744620 MUT:Bam n: Bam Mutant.. | V0892:GSM628272 IP:total RNA MUT:ago2 ago2[414] ovary total RNA | V083:GSM609223 IP:total RNA male, one day | V084:GSM609224 IP:total RNA female, one day | S29:GSM322543 IP:total RNA male head #1 | V007:GSM609247 IP:total RNA heat female head | SRR298711:GSM744619 SETDB1 Mutant.. | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | SRR031696:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | V126: IP:total RNA CME L1 | SRR317114:GSM767598 IP:total RNA om 5-6 days old flies.. | SRR298537:GSM744630 MUT:Vret ret Mutant.. | SRR317115:GSM767599 IP:total RNA old flies.. | S1:GSM286601 IP:total RNA male head | V015: IP:total RNA DreRFHV148h | V006: IP:total RNA r2d2 female: possibly heterozygous | V026:GSM609221 IP:total RNA 1182-4H cell | SRR031704:GSM466497 IP:2'Ome in mutant MUT:r2d2 ies.. | SRR351332:GSM811191 IP:total RNA Nibbler ygenotype: control wild type (w, 5905).. | SRR001338:GSM278707 IP:total RNA IR_non-beta-eliminated | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V008: IP:total RNA S2-DRSC | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | V076:GSM609225 IP:total RNA ML-DmBG3-C2 | V125: IP:total RNA ML-DmD9 | GSM467731: MUT:loqs Dmel_loq_sRNAseq | GSM379064: MUT:Vasa Vasa Mutant | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR032096:GSM442736 IP:ago2 MUT:dcr2 AGO2 IP dcr2 knockdown | GW182004 | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | GW182003 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCT.................................................................... | 21 | 1 | 158.00 | 158.00 | 31.00 | 13.00 | 16.00 | 6.00 | 13.00 | 4.00 | 7.00 | 12.00 | 7.00 | 5.00 | 6.00 | 8.00 | 8.00 | 3.00 | 1.00 | 3.00 | - | 2.00 | 2.00 | - | 2.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTG.................................................................. | 23 | 1 | 142.00 | 142.00 | 28.00 | 21.00 | 11.00 | 18.00 | 6.00 | 12.00 | 7.00 | 2.00 | 7.00 | 8.00 | 1.00 | 2.00 | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | 3.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTT................................................................... | 22 | 1 | 32.00 | 32.00 | 5.00 | 2.00 | 3.00 | 2.00 | 1.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTGt................................................................. | 24 | t | 8.00 | 142.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTGa................................................................. | 24 | a | 5.00 | 142.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGGCCGGTCGAGCAGCGTCTTG.................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTt.................................................................. | 23 | t | 4.00 | 32.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTC..................................................................... | 20 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGT...................................................................... | 19 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGACGCCTTCGGCCGGCGCAAA........................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTa.................................................................. | 23 | a | 2.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTg................................................................... | 22 | g | 1.00 | 158.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTcg.................................................................. | 23 | cg | 1.00 | 158.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTGGGGTTGTTCGTG...................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGGCCACGTGCCCGCAAGccgc........................................................................................................................... | 22 | ccgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCa.................................................................... | 21 | a | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CGCAAAGTTTCAATTTcga.............................................................................................. | 19 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TTCCTATTCTGTGGCACTGT...................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTAAGTTCGATTCCTt.................................... | 16 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GTTCGTGGATTAAGTggta.......................................... | 19 | ggta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGgctt................................................................... | 22 | gctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................CGTCTTGGGGTTGTTCGTGGt.................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTGGGGTTGTTCGTGcgg................................................... | 18 | cgg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCctg.................................................................. | 23 | ctg | 1.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCCGGTCGAGCAGCGTCTTG.................................................................. | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGgct.................................................................... | 21 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTtt................................................................. | 24 | tt | 1.00 | 32.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................CACGTGCCCGCAAGACGCC.......................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGGCCGGTCGAGCAGCGTCT.................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................GGTCGAGCAGCGTCTggg................................................................. | 18 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGGCCGGTCGAGCAGCGTCTTc.................................................................. | 23 | c | 1.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTGGGGTTGTTCGTGGtggc................................................. | 20 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GCGTCTTGGGG............................................................... | 11 | 16 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTTGGGGTTGTT.......................................................... | 12 | 19 | 0.21 | 0.21 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.21 | - | - | - | - |
| ....................................................................................................................................................................TGGCACTGTGGCT.................. | 13 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| ...........................................................................................................................................TGGATTAAGTTC............................................ | 12 | 16 | 0.19 | 0.19 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | 0.06 |
| ...........................................................................................................TGGCCGGTCGAG............................................................................ | 12 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - |
| ..................................................................................................................................................AGTTCGATTCCT..................................... | 12 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCGTCGAAAGGGTAATTGAAGAAGCGAAAACCTGGGTTATCCCTGCCACATGGCCACGTGCCCGCAAGACGCCTTCGGCCGGCGCAAAGTTTCAATTTGATATTCCTTGGCCGGTCGAGCAGCGTCTTGGGGTTGTTCGTGGATTAAGTTCGATTCCTATTCTGTGGCACTGTGGCTTTTCTCGCCGACAGGCTG .............................................................((.((((((((.((((((((((.((((((.((.....)).)))..)))))))))))))..))))))))))................................................................ .............................................................62...................................................................131.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | V137: IP:total RNA Male aged head | V138: IP:total RNA Male cold body | V136: IP:total RNA Male aged body | V141: IP:total RNA Heat_female_body | V142: IP:total RNA Oxidation_female_body | V139: IP:total RNA Cold_female_body | V077: IP:total RNA cold, female head | V080: IP:total RNA Starvation, female head | V140: IP:total RNA Dessication_female_body | V078: IP:total RNA Desiccation, female head | V090:GSM609234 IP:total RNA CS male total RNA | V079: IP:2'Ome in mutant Oxidation, female head | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V086: IP:total RNA female body, aged | S24:GSM399107 IP:total RNA male body #2 | V013:GSM609251 IP:total RNA aged female head | SRR317111:GSM767595 IP:total RNA : from 5-6 days old flies.. | GSM467729: IP:total RNA Dmel_wt_sRNAseq | V098:GSE24544 IP:total RNA MUT:dcr2 dcr-2[L811fsX] ovary total RNA | V132: IP:total RNA ML-DmD32 | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR298712:GSM744620 MUT:Bam n: Bam Mutant.. | V0892:GSM628272 IP:total RNA MUT:ago2 ago2[414] ovary total RNA | V083:GSM609223 IP:total RNA male, one day | V084:GSM609224 IP:total RNA female, one day | S29:GSM322543 IP:total RNA male head #1 | V007:GSM609247 IP:total RNA heat female head | SRR298711:GSM744619 SETDB1 Mutant.. | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | SRR031696:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | V126: IP:total RNA CME L1 | SRR317114:GSM767598 IP:total RNA om 5-6 days old flies.. | SRR298537:GSM744630 MUT:Vret ret Mutant.. | SRR317115:GSM767599 IP:total RNA old flies.. | S1:GSM286601 IP:total RNA male head | V015: IP:total RNA DreRFHV148h | V006: IP:total RNA r2d2 female: possibly heterozygous | V026:GSM609221 IP:total RNA 1182-4H cell | SRR031704:GSM466497 IP:2'Ome in mutant MUT:r2d2 ies.. | SRR351332:GSM811191 IP:total RNA Nibbler ygenotype: control wild type (w, 5905).. | SRR001338:GSM278707 IP:total RNA IR_non-beta-eliminated | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V008: IP:total RNA S2-DRSC | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | V076:GSM609225 IP:total RNA ML-DmBG3-C2 | V125: IP:total RNA ML-DmD9 | GSM467731: MUT:loqs Dmel_loq_sRNAseq | GSM379064: MUT:Vasa Vasa Mutant | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR032096:GSM442736 IP:ago2 MUT:dcr2 AGO2 IP dcr2 knockdown | GW182004 | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | GW182003 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TGGGGTTGTTCG........................................................ | 12 | 12 | 1.58 | 1.58 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.92 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | 0.08 | - |
| .................................................................gcccCGCCTTCGGCCGGCG............................................................................................................... | 19 | gccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................attAGTTCGATTCCTATTCT................................ | 20 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................cCAGCGTCTTGGGGTTGTTCGTGGA.................................................... | 25 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................GCGTCTTGGGGTTGTTCGTGGATTAAG............................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGGGGTTGTTCGT....................................................... | 13 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GCCTTCGGCCGG................................................................................................................. | 12 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCCGGTCGAG............................................................................ | 11 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGACGCCTT........................................................................................................................ | 11 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |