|
(2) 1182-4H |
(1) 2OME.ip |
(1) 2OME_MUTANT.ip |
(4) AGO1.ip |
(1) AGO3.ip |
(1) BODY |
(2) CMEW1 Cl.8+ cell |
(2) DCR2.mut |
(1) EMBRYO |
(6) HEAD |
(1) KC |
(4) ML_CELL_LINE |
(1) OSS |
(1) OTHER.mut |
(14) OVARY |
(4) R2D2.mut |
(2) S2 |
(1) S2_NP |
(1) S2_R |
(1) TESTES |
(34) TOTAL_RNA.ip |
(3) WHOLE |
(1) WING |
(1) mbn2 |
| TATGCTTCATATTGTTAAGAAATCAACGAAAACTTTGAGCGTTAAAAAGAAGTTTCCTTACTTCAAATTCTTGTTGCATTTCCAACGCTTTCAGAATTTGAAACTTTGGAAATGTAACATCTTTAAAGTTGATGGGAGTACACATGCAAAAATATTGCGGGATCATTAAAAATACGAAAAGGTATTTTTCAGACC ...................................................(((((((.((((.(((....((((((((((((((...((((((...))))))..))))))))))))))..))).))))....))))).))...................................................... ..................................................51............................................................................................145................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | V073: IP:total RNA mbn2 | SRR029028:GSM430030 IP:total RNA untreated (mock) | SRR014278:GSM327632 IP:total RNA Ovary_rep1_w1118(fem)_x_Har(male)_F1 | V085: IP:total RNA CME W2 wing disc | V022: IP:total RNA ML-DmD32 cell | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | V086: IP:total RNA female body, aged | SRR060644:GSM548583 IP:total RNA A2_ovaries_total | SRR317112:GSM767596 IP:total RNA old flies.. | V083:GSM609223 IP:total RNA male, one day | SRR014275:GSM327624 IP:total RNA Ovary_rep1_LK_P | V097:GSE24545 IP:total RNA CS ovary total RNA | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V0652: IP:ago1 MUT:dcr2 dcr2 ovary, AGO1-IP, reseq | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | S32:GSM609242 IP:total RNA s2+48 #2 | SRR097867:GSM666013 IP:total RNA Drosophila S2-NP cells | SRR298537:GSM744630 MUT:Vret ret Mutant.. | SRR010954:SRX002239 IP:2'Ome in mutant Aub trans-heterozygotes, oxidized | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR060643:GSM548582 IP:total RNA A2_testes_total | V095:SRX247212 IP:ago1 AGO1-IP from S2-R+, FLAG-HA-AGO2 | SRR010959:SRX002244 IP:ago3 Ago3 IP in heterozygotes | V026:GSM609221 IP:total RNA 1182-4H cell | V075:GSM609222 IP:total RNA ML-DmBG1-C1 | V147: IP:total RNA 1182-4H cell | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V084:GSM609224 IP:total RNA female, one day | SRR010953:SRX002238 IP:total RNA Aub heterozygotes, oxidized | V088:GSM609232 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | GSM379056: IP:total RNA Krimp Heterozygote | V076:GSM609225 IP:total RNA ML-DmBG3-C2 | SRR014269:GSM327629 IP:total RNA Embryo_0-2hrs_rep1_LK_0_2hr_Emb | SRR031698:GSM466491 IP:2Ome flies.. | V028:GSM609226 IP:total RNA CMEW1 Cl.8+ cell | V035:GSM609250 IP:total RNA ML-DmD32 cell | V091: IP:total RNA fGS/OSS total | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V024:GSM399101 IP:total RNA kc167 cell | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................ACTTTGGAAATGTAACATCTTT....................................................................... | 22 | 1 | 6.00 | 6.00 | - | 3.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........TATTGTTAAGAAATCtttt....................................................................................................................................................................... | 19 | tttt | 5.00 | 0.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - |
| .........TATTGTTAAGAAATCttt........................................................................................................................................................................ | 18 | ttt | 4.00 | 0.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AACTTTGGAAATGTAACATCTT........................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................AAATACGAAAAGGTAaaa........ | 18 | aaa | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................AAACTTTGGAAATGTAACATCT......................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................AAACTTTGGAAATGTAACATC.......................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTGTTGCATTTCCAACGatga........................................................................................................ | 21 | atga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ACTTTGGAAATGTAACATCTTTA...................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................ATTAAAAATACGAAAAGGTATTTT....... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................AAATACGAAAAGGTATagg....... | 19 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TTCATATTGTTAAGAttt............................................................................................................................................................................ | 18 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGTAACATCTTTAAAtggg................................................................ | 19 | tggg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ATGTAACATCTTTAAAGcaga............................................................... | 21 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TTTCCTTACTTCAAAgcg............................................................................................................................. | 18 | gcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TAAAAAGAAGTTTCCTTgaag.................................................................................................................................... | 21 | gaag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ACTTTGGAAATGTAACATCT......................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ACTTTGGAAATGTAACATCTTa....................................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GAAGTTTCCTTACTTac.................................................................................................................................. | 17 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................GTTTCCTTACTTC................................................................................................................................... | 13 | 7 | 0.43 | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.43 |
| .................................................AAGTTTCCTTACTT.................................................................................................................................... | 14 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TTTCAGAATTTGA.............................................................................................. | 13 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
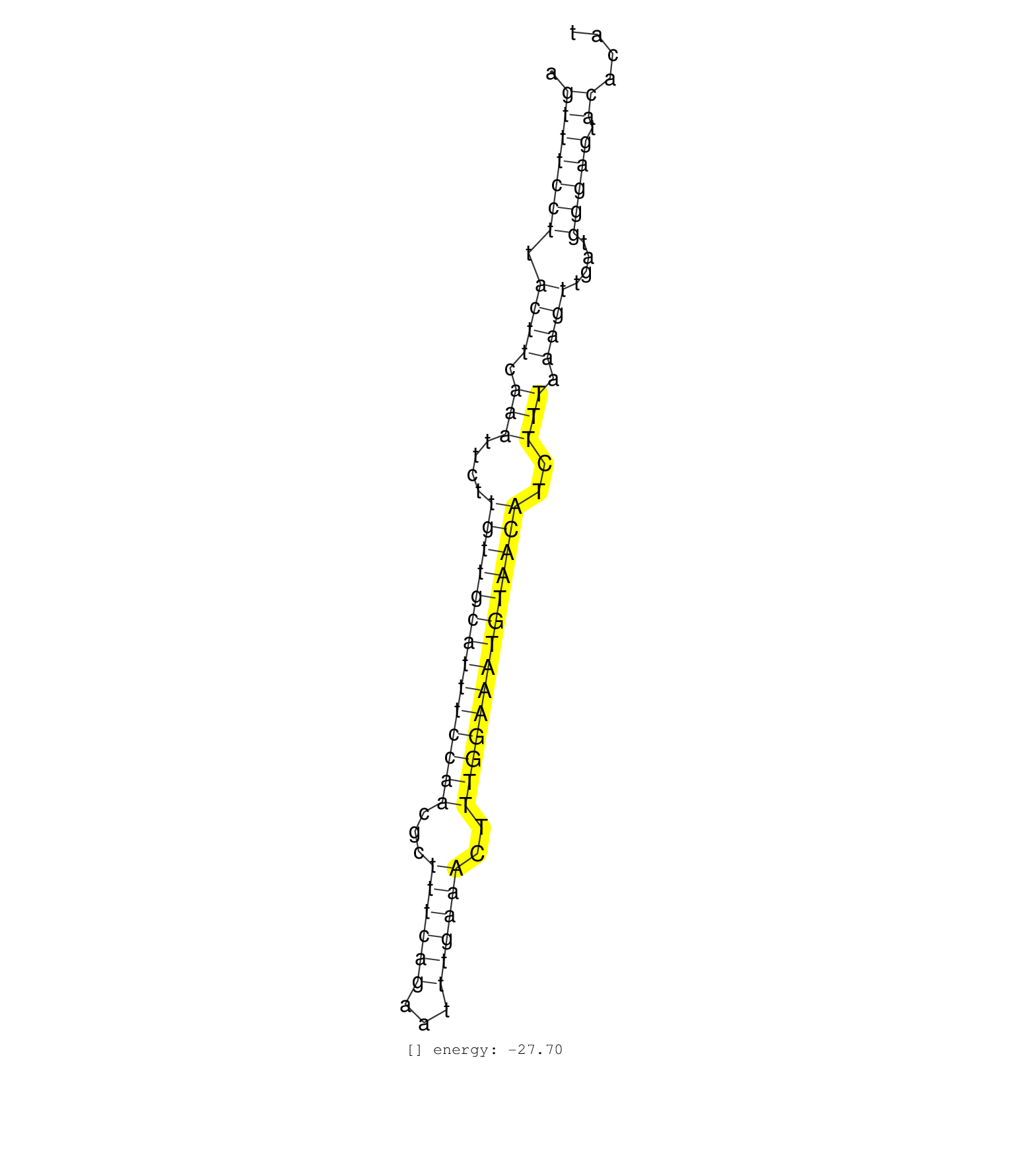
| TATGCTTCATATTGTTAAGAAATCAACGAAAACTTTGAGCGTTAAAAAGAAGTTTCCTTACTTCAAATTCTTGTTGCATTTCCAACGCTTTCAGAATTTGAAACTTTGGAAATGTAACATCTTTAAAGTTGATGGGAGTACACATGCAAAAATATTGCGGGATCATTAAAAATACGAAAAGGTATTTTTCAGACC ...................................................(((((((.((((.(((....((((((((((((((...((((((...))))))..))))))))))))))..))).))))....))))).))...................................................... ..................................................51............................................................................................145................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | V073: IP:total RNA mbn2 | SRR029028:GSM430030 IP:total RNA untreated (mock) | SRR014278:GSM327632 IP:total RNA Ovary_rep1_w1118(fem)_x_Har(male)_F1 | V085: IP:total RNA CME W2 wing disc | V022: IP:total RNA ML-DmD32 cell | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V0662: IP:ago1 MUT:r2d2 r2d2 ovary, AGO1-IP, reseq | V086: IP:total RNA female body, aged | SRR060644:GSM548583 IP:total RNA A2_ovaries_total | SRR317112:GSM767596 IP:total RNA old flies.. | V083:GSM609223 IP:total RNA male, one day | SRR014275:GSM327624 IP:total RNA Ovary_rep1_LK_P | V097:GSE24545 IP:total RNA CS ovary total RNA | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | V0652: IP:ago1 MUT:dcr2 dcr2 ovary, AGO1-IP, reseq | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | S32:GSM609242 IP:total RNA s2+48 #2 | SRR097867:GSM666013 IP:total RNA Drosophila S2-NP cells | SRR298537:GSM744630 MUT:Vret ret Mutant.. | SRR010954:SRX002239 IP:2'Ome in mutant Aub trans-heterozygotes, oxidized | SRR031703:GSM466496 IP:total RNA MUT:r2d2 Total small RNAs from r2d2 homozygous flies | SRR060643:GSM548582 IP:total RNA A2_testes_total | V095:SRX247212 IP:ago1 AGO1-IP from S2-R+, FLAG-HA-AGO2 | SRR010959:SRX002244 IP:ago3 Ago3 IP in heterozygotes | V026:GSM609221 IP:total RNA 1182-4H cell | V075:GSM609222 IP:total RNA ML-DmBG1-C1 | V147: IP:total RNA 1182-4H cell | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | V033:GSM609227 IP:total RNA CMEW1 Cl.8+ cell | V084:GSM609224 IP:total RNA female, one day | SRR010953:SRX002238 IP:total RNA Aub heterozygotes, oxidized | V088:GSM609232 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | GSM379056: IP:total RNA Krimp Heterozygote | V076:GSM609225 IP:total RNA ML-DmBG3-C2 | SRR014269:GSM327629 IP:total RNA Embryo_0-2hrs_rep1_LK_0_2hr_Emb | SRR031698:GSM466491 IP:2Ome flies.. | V028:GSM609226 IP:total RNA CMEW1 Cl.8+ cell | V035:GSM609250 IP:total RNA ML-DmD32 cell | V091: IP:total RNA fGS/OSS total | SRR032152:GSM466489 IP:ago1 Ago1-associated small RNAs from Oregon R | V024:GSM399101 IP:total RNA kc167 cell | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................aaaGCATTTCCAACGCTTT........................................................................................................ | 19 | aaa | 14.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | 2.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................cacTGTTGCATTTCCAAC............................................................................................................. | 18 | cac | 5.00 | 0.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................ccccCACATGCAAAAATATT....................................... | 20 | cccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................cgcTGTTGCATTTCCAAC............................................................................................................. | 18 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................cagTGTTGCATTTCCAAC............................................................................................................. | 18 | cag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................gggTGTTGCATTTCCAAC............................................................................................................. | 18 | ggg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................tctCATGCAAAAATATTG...................................... | 18 | tct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........aggcTGTTAAGAAATCAAC........................................................................................................................................................................ | 19 | aggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................cccTGTTGCATTTCCAAC............................................................................................................. | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................aaaaCATTTCCAACGCTTT........................................................................................................ | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ccctCACATGCAAAAATAT........................................ | 19 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................aaGCATTTCCAACGCTTT........................................................................................................ | 18 | aa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................cggcAATCAACGAAAACTT................................................................................................................................................................ | 19 | cggc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................cccCACATGCAAAAATAT........................................ | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |