|
(1) 2OME.ip |
(1) 2OME_MUTANT.ip |
(2) AGO1.ip |
(3) AGO2.mut |
(4) BODY |
(2) DCR2.mut |
(2) EMBRYO |
(1) GM2 |
(9) HEAD |
(1) OTHER.ip |
(3) OTHER.mut |
(2) OVARY |
(2) S2 |
(2) S2_R |
(2) TESTES |
(24) TOTAL_RNA.ip |
(4) WHOLE |
(1) WING |
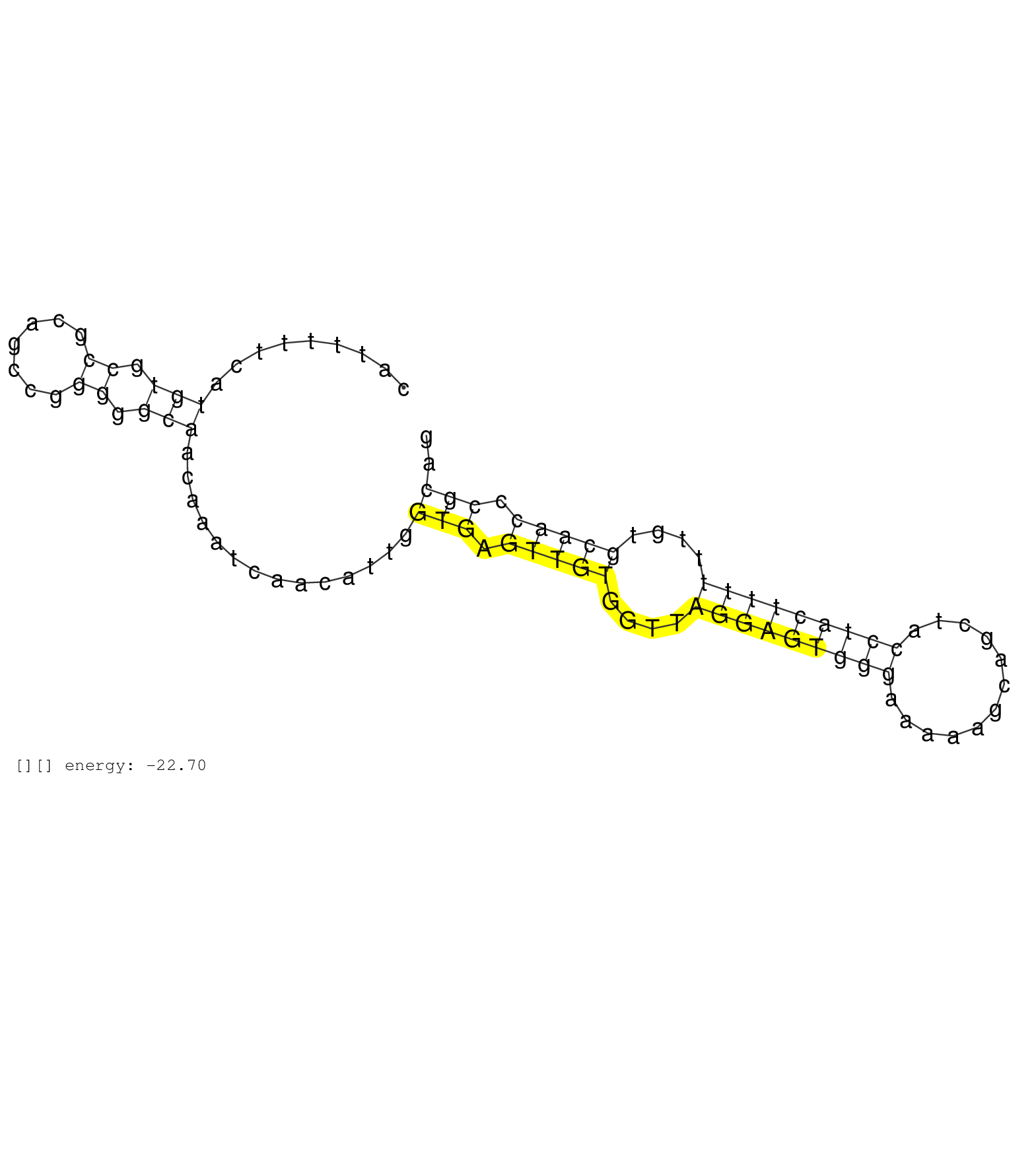
| AGCGCATACTTTTGCTCGGCTGCATGTGGAGTGCATTTTTCATGTGCCGCAGCCGGGGGCAACAAATCAACATTGGTGAGTTGTGGTTAGGAGTGGGAAAAAGCAGCTACCTACTTTTTTGTGCAACCCGCAGGCGCCTTCTTCTACGACGATGAGCTGGAGCTGGAGAAAGAGTTCATGACGGTGGTCAATGCCATCAATGGACCTG ..........................................(((.((.......)).)))..............(((.(((((....(((((((((............)))))))))....))))).)))............................................................................. .................................34.................................................................................................133......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | GSM467729: IP:total RNA Dmel_wt_sRNAseq | SRR060645:GSM548584 IP:total RNA yw67c23(2)_testes_total | V086: IP:total RNA female body, aged | V138: IP:total RNA Male cold body | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | S24:GSM399107 IP:total RNA male body #2 | V019:GSM609219 IP:total RNA GM2 cell | GSM379065: IP:total RNA Zuc Heterozygote | S32:GSM609242 IP:total RNA s2+48 #2 | V085: IP:total RNA CME W2 wing disc | V137: IP:total RNA Male aged head | V083:GSM609223 IP:total RNA male, one day | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | SRR060652:GSM548591 IP:total RNA hs-Penelope_testes_total | SRR014269:GSM327629 IP:total RNA Embryo_0-2hrs_rep1_LK_0_2hr_Emb | SRR341116:GSM786605 IP:Luciferase MUT:Oxidized pe: Schneider 2 (S2) cellsoxidation.. | V077: IP:total RNA cold, female head | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR031698:GSM466491 IP:2Ome flies.. | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | V095:SRX247212 IP:ago1 AGO1-IP from S2-R+, FLAG-HA-AGO2 | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | V064:GSM609231 IP:ago1 MUT:ago2 ago2[414], ovary, AGO1IP | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................ATGTGGAGTGCATTTTTCAc..................................................................................................................................................................... | 20 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................CAGCCGGGGGCAACAAATCAACAT....................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGAGTTGTGGTTAGGtgt.................................................................................................................. | 18 | tgt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGGAGTGGGAAAAAGgctt..................................................................................................... | 19 | gctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TTCATGACGGTGGTCAATGCC............. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGGAGTGGGAAAAAGaa....................................................................................................... | 17 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GTGAGTTGTGGTTAGaac................................................................................................................... | 18 | aac | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GTGAGTTGTGGTTAGGAGT.................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GGTGAGTTGTGGTTAcgg.................................................................................................................... | 18 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CGGCTGCATGTGGAGTGCATT........................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GCTGGAGCTGGAGAAAGAGTTCA.............................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ATGTGGAGTGCATTTTTCAca.................................................................................................................................................................... | 21 | ca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AGCTGGAGCTGGAGAtgca................................... | 19 | tgca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TTGTGGTTAGGAGTGaaa.............................................................................................................. | 18 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................TGAGTTGTGGTTAGGggt.................................................................................................................. | 18 | ggt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACTTTTTTGTGCAAgca............................................................................... | 18 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GCTGGAGCTGGAGAAggg................................... | 18 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GTGAGTTGTGGTTAGGAGTG................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GAGTTCATGACG......................... | 12 | 7 | 0.43 | 0.43 | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - |
| .................................................................................TGTGGTTAGGAGT.................................................................................................................. | 13 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGCAGCCGGGGGCA................................................................................................................................................... | 14 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| ..............................................................................AGTTGTGGTTAGGA.................................................................................................................... | 14 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - |
| ..................................................AGCCGGGGGCAA.................................................................................................................................................. | 12 | 19 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | 0.05 | - | - | - |
| ...............................................................................GTTGTGGTTAGG..................................................................................................................... | 12 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - |
| .............................................................................................................................ACCCGCAGGCGC....................................................................... | 12 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 | - | - | - |
| .......................................................................................................................................GCCTTCTTCTAC............................................................. | 12 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
| AGCGCATACTTTTGCTCGGCTGCATGTGGAGTGCATTTTTCATGTGCCGCAGCCGGGGGCAACAAATCAACATTGGTGAGTTGTGGTTAGGAGTGGGAAAAAGCAGCTACCTACTTTTTTGTGCAACCCGCAGGCGCCTTCTTCTACGACGATGAGCTGGAGCTGGAGAAAGAGTTCATGACGGTGGTCAATGCCATCAATGGACCTG ..........................................(((.((.......)).)))..............(((.(((((....(((((((((............)))))))))....))))).)))............................................................................. .................................34.................................................................................................133......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | GSM467729: IP:total RNA Dmel_wt_sRNAseq | SRR060645:GSM548584 IP:total RNA yw67c23(2)_testes_total | V086: IP:total RNA female body, aged | V138: IP:total RNA Male cold body | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | S24:GSM399107 IP:total RNA male body #2 | V019:GSM609219 IP:total RNA GM2 cell | GSM379065: IP:total RNA Zuc Heterozygote | S32:GSM609242 IP:total RNA s2+48 #2 | V085: IP:total RNA CME W2 wing disc | V137: IP:total RNA Male aged head | V083:GSM609223 IP:total RNA male, one day | SRR031701:GSM466494 IP:total RNA Total small RNAs from r2d2 heterozygous flies | V092: IP:total RNA MUT:dcr2 dcr-2[G31R] male total RNA | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | V012: IP:total RNA Dcr2 male (Katsutomo, whole fly?) | SRR060652:GSM548591 IP:total RNA hs-Penelope_testes_total | SRR014269:GSM327629 IP:total RNA Embryo_0-2hrs_rep1_LK_0_2hr_Emb | SRR341116:GSM786605 IP:Luciferase MUT:Oxidized pe: Schneider 2 (S2) cellsoxidation.. | V077: IP:total RNA cold, female head | V081:GSM609229 IP:total RNA embryo 2-6hr | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | SRR031698:GSM466491 IP:2Ome flies.. | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | V095:SRX247212 IP:ago1 AGO1-IP from S2-R+, FLAG-HA-AGO2 | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | V064:GSM609231 IP:ago1 MUT:ago2 ago2[414], ovary, AGO1IP | SRR351333:GSM811192 IP:total RNA MUT:Nibbler ygenotype: nbr[f02257].. | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................ggCTTCTTCTACGACGATG...................................................... | 19 | gg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................gcccTTCATGACGGTGGTCA.................. | 20 | gccc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................gggCTTCTTCTACGACGATG...................................................... | 20 | ggg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GAGTTGTGGTTAGG..................................................................................................................... | 14 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................tgGTGCCGCAGCCGGGG...................................................................................................................................................... | 17 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ggCTTCTTCTACGACGATGAG.................................................... | 21 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................gggCTTCTTCTACGACGATGA..................................................... | 21 | ggg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGCAACCCGCAG........................................................................... | 12 | 7 | 0.29 | 0.29 | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - |
| ........................................................................................................................GTGCAACCCGCA............................................................................ | 12 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| .............................................GCCGCAGCCGGGG...................................................................................................................................................... | 13 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ............................................TGCCGCAGCCGGG....................................................................................................................................................... | 13 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| ............................................................................................................................AACCCGCAGGCG........................................................................ | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - |
| ...........................................................................................................................CAACCCGCAGGC......................................................................... | 12 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - |
| .............................................GCCGCAGCCGGG....................................................................................................................................................... | 12 | 11 | 0.09 | 0.09 | - | - | - | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GTGCAACCCGC............................................................................. | 11 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - |