|
(1) 2OME.ip |
(3) 2OME_MUTANT.ip |
(1) AGO1.ip |
(2) AGO2.ip |
(3) AGO2.mut |
(4) BODY |
(3) DCR2.mut |
(2) DISC |
(1) EMBRYO |
(10) HEAD |
(2) LOQS.mut |
(1) ML_CELL_LINE |
(1) OSS |
(4) OTHER.mut |
(8) OVARY |
(3) PIWI.ip |
(2) R2D2.mut |
(1) S2 |
(1) S2_NP |
(4) S2_R |
(1) S3 |
(3) TESTES |
(28) TOTAL_RNA.ip |
(2) WHOLE |
(1) WING |
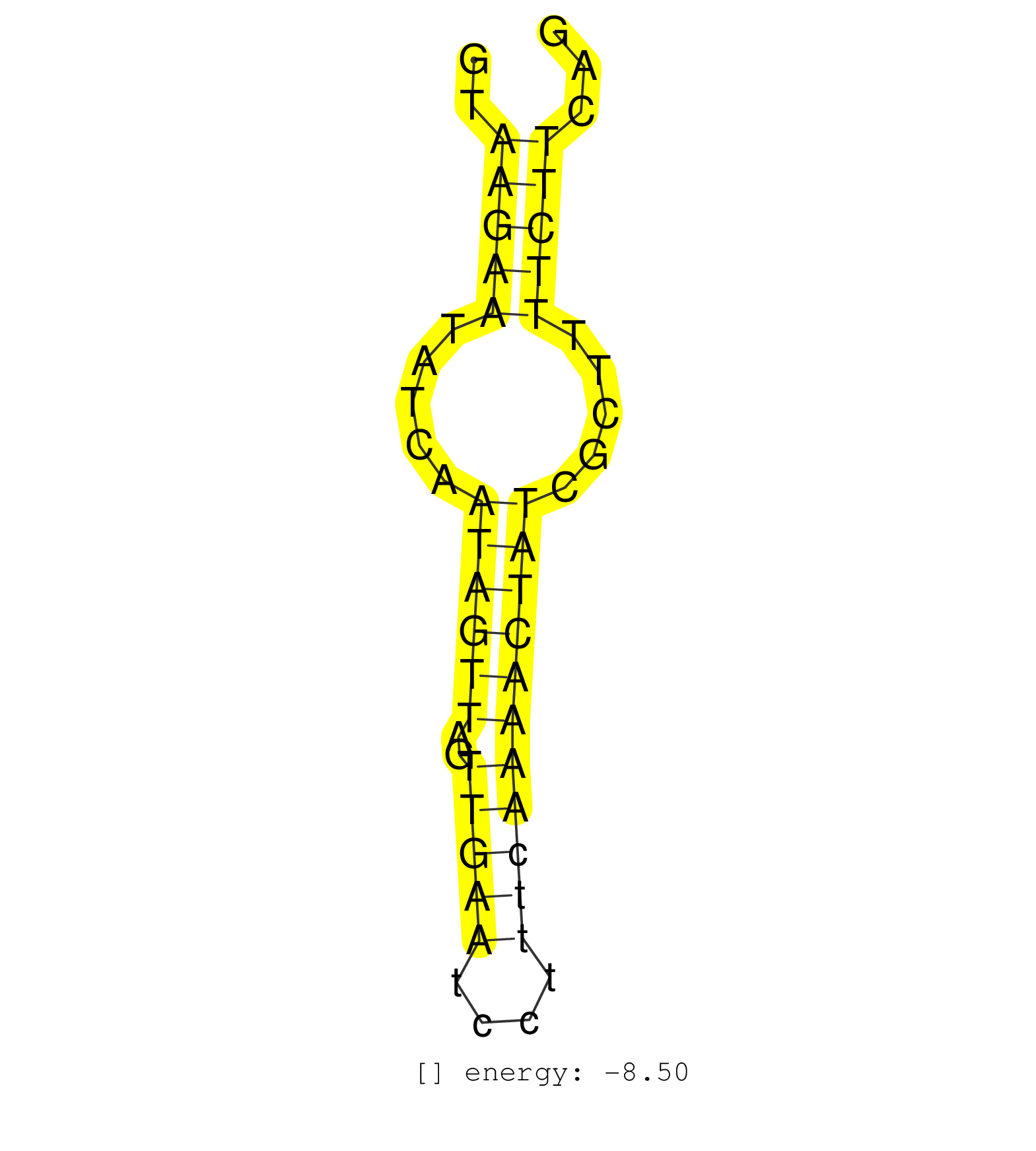
| TTGAGAATATAAATTTGGGAAAAGAACTGAAGGTCCGCGCTTCGCAGCAGTTTCAACTTGCGCTGAATTTGATTGGTAAGAATATCAATAGTTAGTTGAATCCTTTCAAAACTATCGCTTTTCTTCAGATGATCACTACTTTTATAGTTTGCTAATTTTTATGGCCACCTTGGCGTACGTGGTCTATATCCTGTGCTACGATA .............................................................................(((((.....((((((..(((((....))))))))))).....))))).............................................................................. ...........................................................................76..................................................128......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR032092:GSM442732 IP:total RNA mock oxidized | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | SRR060645:GSM548584 IP:total RNA yw67c23(2)_testes_total | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V067:GSM371638 IP:total RNA S2-NP | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | GSM467731: MUT:loqs Dmel_loq_sRNAseq | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR060644:GSM548583 IP:total RNA A2_ovaries_total | SRR065152:GSM577959 IP:piwi .. | SRR065155:GSM577962 IP:piwi ies.. | SRR001340:GSM278575 IP:2Ome IR_beta-eliminated | V085: IP:total RNA CME W2 wing disc | V015: IP:total RNA DreRFHV148h | S5:GSM275691 IP:total RNA imaginal disc | V075:GSM609222 IP:total RNA ML-DmBG1-C1 | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | GSM343287: IP:total RNA Drosophila Toll 10b mutant embryos | M067: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | V084:GSM609224 IP:total RNA female, one day | SRR010953:SRX002238 IP:total RNA Aub heterozygotes, oxidized | V074: IP:total RNA S3 | M068: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR060652:GSM548591 IP:total RNA hs-Penelope_testes_total | SRR065153:GSM577960 IP:piwi .. | M065: MUT:CG1091 ovary - sample C, homozygous mutant | V091: IP:total RNA fGS/OSS total | S21:GSM399105 IP:total RNA disk #2 | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | V086: IP:total RNA female body, aged | GSM280085:GSM280085 IP:total RNA WT testes (18-24nt) | V138: IP:total RNA Male cold body | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR031700:GSM466493 IP:2'Ome in mutant MUT:dcr2 lies.. | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | SRR001344:GSM278692 IP:2'Ome in mutant MUT:dcr2 dcr-2_beta-eliminated | V064:GSM609231 IP:ago1 MUT:ago2 ago2[414], ovary, AGO1IP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TACGTGGTCTATATCtgg.......... | 18 | tgg | 10.00 | 0.00 | - | 2.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TACGTGGTCTATATCggg.......... | 18 | ggg | 8.00 | 0.00 | - | 5.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AAAACTATCGCTTTTCTTCAGt.......................................................................... | 22 | t | 6.00 | 1.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...AGAATATAAATTTGGGAAAA.................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATCGCTTTTCTTCAGAgtt....................................................................... | 19 | gtt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AAAACTATCGCTTTTCTTCAG........................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................TTTGATTGGTAAGAAgcta..................................................................................................................... | 19 | gcta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........AAATTTGGGAAAAGAAagg.............................................................................................................................................................................. | 19 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................GCTGAATTTGATTGGc.............................................................................................................................. | 16 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GAATTTGATTGGTAAaag......................................................................................................................... | 18 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GGTCTATATCCTGTGCTA..... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................ACTTGCGCTGAATTT..................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GTAAGAATATCAATAGTTAGTTGAA....................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................CTATATCCTGTGCTAg.... | 16 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CGCTGAATTTGATTGcga............................................................................................................................. | 18 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCTGAATTTGATTGGga............................................................................................................................. | 17 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGGGAAAAGAACTGAggag......................................................................................................................................................................... | 19 | ggag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GCAGCAGTTTCAACTTcga............................................................................................................................................. | 19 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................GCTGAATTTGATTGGacct........................................................................................................................... | 19 | acct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AAAGAACTGAAGGTCgtg..................................................................................................................................................................... | 18 | gtg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCTGAATTTGATTGGatta........................................................................................................................... | 19 | atta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TACGTGGTCTATATCttt.......... | 18 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GGTAAGAATATCAATgagc.............................................................................................................. | 19 | gagc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AAAACTATCGCTTTTCTTCAGc.......................................................................... | 22 | c | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGTTAGTTGAATC..................................................................................................... | 13 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| .....................................................................................................................................................................CACCTTGGCGTA.......................... | 12 | 18 | 0.06 | 0.06 | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTGAGAATATAAATTTGGGAAAAGAACTGAAGGTCCGCGCTTCGCAGCAGTTTCAACTTGCGCTGAATTTGATTGGTAAGAATATCAATAGTTAGTTGAATCCTTTCAAAACTATCGCTTTTCTTCAGATGATCACTACTTTTATAGTTTGCTAATTTTTATGGCCACCTTGGCGTACGTGGTCTATATCCTGTGCTACGATA .............................................................................(((((.....((((((..(((((....))))))))))).....))))).............................................................................. ...........................................................................76..................................................128......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR032092:GSM442732 IP:total RNA mock oxidized | SRR029608: IP:total RNA total small RNAs from hen1 heterozygous flies | SRR029633: IP:total RNA total small RNAs from hen1 homozygous flies | SRR001347:GSM278704 IP:total RNA MUT:ago2 ago2_untreated | V093: IP:total RNA MUT:loqs loqs-KO, 40A; loqs-PB rescue/TM3 male total RNA | SRR060645:GSM548584 IP:total RNA yw67c23(2)_testes_total | SRR031692:GSM466487 IP:total RNA Total small RNAs from Oregon R | V067:GSM371638 IP:total RNA S2-NP | M069: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR001349:GSM278705 IP:total RNA heterozygous_dcr-2_untreated | GSM467731: MUT:loqs Dmel_loq_sRNAseq | SRR001348:GSM278693 IP:2'Ome in mutant MUT:ago2 ago2_oxidized | SRR060644:GSM548583 IP:total RNA A2_ovaries_total | SRR065152:GSM577959 IP:piwi .. | SRR065155:GSM577962 IP:piwi ies.. | SRR001340:GSM278575 IP:2Ome IR_beta-eliminated | V085: IP:total RNA CME W2 wing disc | V015: IP:total RNA DreRFHV148h | S5:GSM275691 IP:total RNA imaginal disc | V075:GSM609222 IP:total RNA ML-DmBG1-C1 | SRR001664:GSM278703 IP:total RNA MUT:dcr2 homozygous_dcr-2_untreated | GSM343287: IP:total RNA Drosophila Toll 10b mutant embryos | M067: IP:total RNA MUT:CG1091 Flag-HA-Ago2 S2R+, knockdown DS1091 | V084:GSM609224 IP:total RNA female, one day | SRR010953:SRX002238 IP:total RNA Aub heterozygotes, oxidized | V074: IP:total RNA S3 | M068: IP:AGO2 MUT:CG1091 Flag-HA-Ago2 S2R+, overexpression UAS1091 | SRR060652:GSM548591 IP:total RNA hs-Penelope_testes_total | SRR065153:GSM577960 IP:piwi .. | M065: MUT:CG1091 ovary - sample C, homozygous mutant | V091: IP:total RNA fGS/OSS total | S21:GSM399105 IP:total RNA disk #2 | GSM467730: MUT:r2d2 Dmel_r2d2_sRNAseq | V086: IP:total RNA female body, aged | GSM280085:GSM280085 IP:total RNA WT testes (18-24nt) | V138: IP:total RNA Male cold body | V094:SRX247213 IP:ago2 FLAG-IP from S2-R+, FLAG-HA-AGO2 | SRR031700:GSM466493 IP:2'Ome in mutant MUT:dcr2 lies.. | V0882:GSM628271 IP:total RNA MUT:r2d2 r2d2[1] ovary total RNA   | SRR001344:GSM278692 IP:2'Ome in mutant MUT:dcr2 dcr-2_beta-eliminated | V064:GSM609231 IP:ago1 MUT:ago2 ago2[414], ovary, AGO1IP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................agcGGCCACCTTGGCGTAC......................... | 19 | agc | 11.00 | 0.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................agttCCACCTTGGCGTACG........................ | 19 | agtt | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................aacaAATCCTTTCAAAACTA......................................................................................... | 20 | aaca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................tcgTAGTTGAATCCTTTCA............................................................................................... | 19 | tcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................ccctTCCTTTCAAAACTAT........................................................................................ | 19 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................gcgTTCAAAACTATCGCT.................................................................................... | 18 | gcg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................cggcTTTCAACTTGCGCTGA......................................................................................................................................... | 20 | cggc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................atctTTTTCTTCAGATGATC..................................................................... | 20 | atct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TCTTCAGATGATCACTACT............................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................aacaTTGAATCCTTTCAAAA............................................................................................ | 20 | aaca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GGTAAGAATATCAATAG................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................tcagCTTTCAAAACTATCGCT.................................................................................... | 21 | tcag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCTGAATTTGATTG................................................................................................................................ | 14 | 8 | 0.62 | 0.62 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | 0.12 | - |
| ..............................................................CTGAATTTGATTG................................................................................................................................ | 13 | 17 | 0.35 | 0.35 | - | - | - | - | - | - | - | - | - | - | - | 0.35 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ACTTGCGCTGAA........................................................................................................................................ | 12 | 9 | 0.22 | 0.22 | - | - | - | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TTTCAACTTGCG............................................................................................................................................. | 12 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| ...................................................................................................................................ATCACTACTTTT............................................................ | 12 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................AGTTGAATCCTT.................................................................................................. | 12 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |