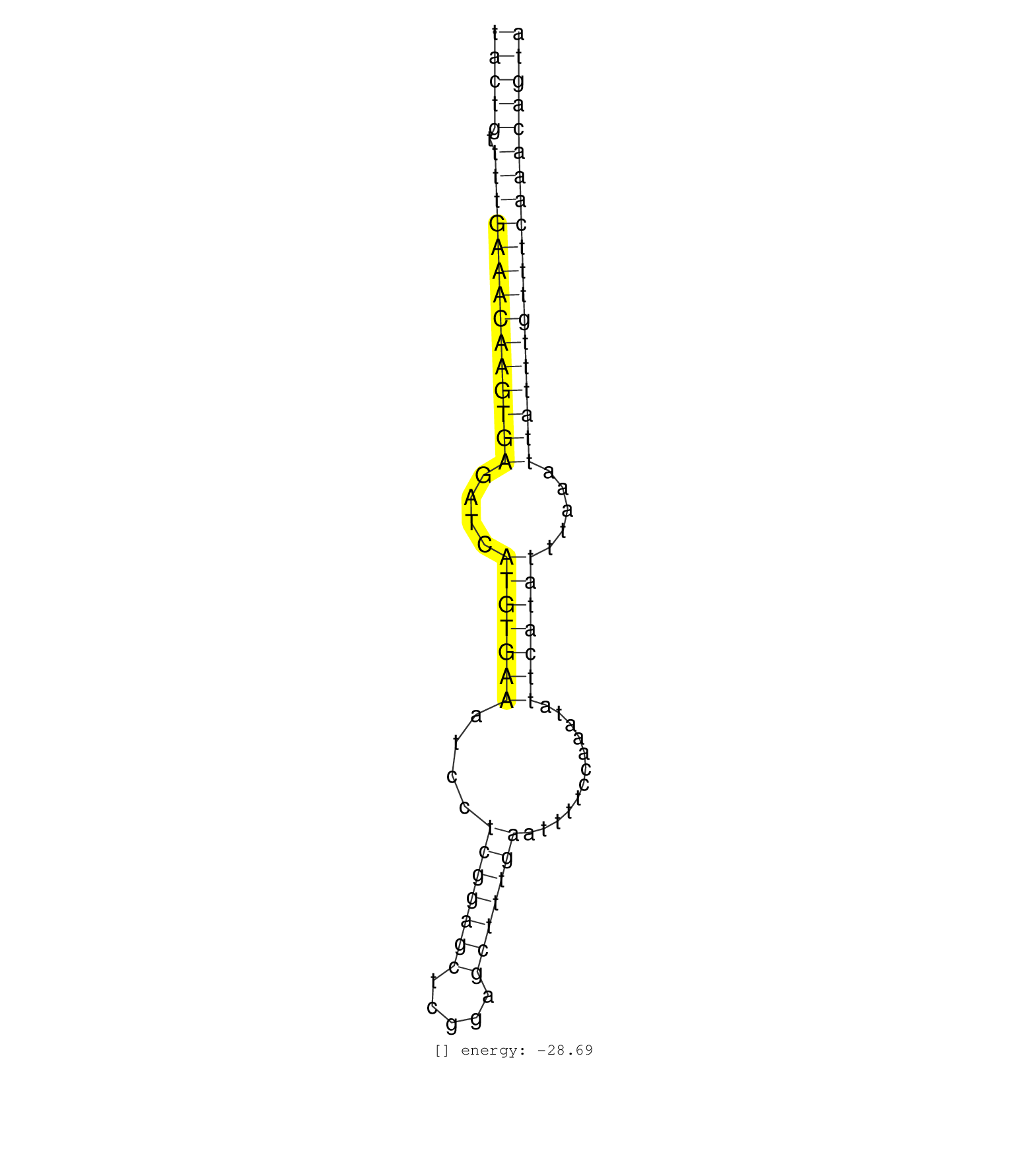
chrIV:16472994-16473190 -
score=-28.69
score=-28.69

| ID: cel-mir-1021 |
 chrIV:16472994-16473190 - score=-28.69 |
 |
| GTTTATGAAATGCAAAAAAGATTTTTCCAGTGTTCCAACTATTTCAGAAATACTGTTTTGAAACAAGTGAGATCATGTGAAATCCTCGGAGCTCGGAGCTTTGAATTTTCCAAATATTCATATTTAAATTATTTGTTTCAAACAGTACGAACTGAAAAATTTTCTGTTTCAGAATGTCAATTTGTAGACATGCACTT ..................................................(((((.((((((((((((((....(((((((....(((((((.....)))))))............))))))).....))))))))))))))))))).................................................. ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM455402 | GSM455401 | GSM455395 | GSM455392 | GSM642429 | GSM455398 | GSM454003 | GSM443999 | GSM455404 | GSM632210 | GSM464607 | GSM444001 | GSM608720 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................GAAACAAGTGAGATCATGTGAA.................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| .......................................................................ATCATGTGAAATCCTCGGAGCTCT...................................................................................................... | 24 | T | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................................................................................................AAAAATTTTCTGTTTCAGAACT..................... | 22 | CT | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..........................................................................ATGTGAAATCCTCGGAGCTCGGAGCTTTGA............................................................................................. | 30 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..........................................TTCAGAAATACTGTTTTGTTA...................................................................................................................................... | 21 | TTA | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ............................................................AAACAAGTGAGATCATGTGAA.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..........................................TTCAGAAATACTGTTTTGAA....................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ........................................................................TCATGTGAAATCCTCGGAG.......................................................................................................... | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .........ATGCAAAAAAGATTTTTCCAG....................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GTTTATGAAATGCAAAAAAGATTTTTCCAGTGTTCCAACTATTTCAGAAATACTGTTTTGAAACAAGTGAGATCATGTGAAATCCTCGGAGCTCGGAGCTTTGAATTTTCCAAATATTCATATTTAAATTATTTGTTTCAAACAGTACGAACTGAAAAATTTTCTGTTTCAGAATGTCAATTTGTAGACATGCACTT ..................................................(((((.((((((((((((((....(((((((....(((((((.....)))))))............))))))).....))))))))))))))))))).................................................. ..................................................51..............................................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM455402 | GSM455401 | GSM455395 | GSM455392 | GSM642429 | GSM455398 | GSM454003 | GSM443999 | GSM455404 | GSM632210 | GSM464607 | GSM444001 | GSM608720 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................TCTGAAATACTGTTTTGAAAC..................................................................................................................................... | 21 | TCT | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| .........................................................................................................................ATTTAAATTATTTGTTTC.......................................................... | 18 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................................................................GTTTCAAACAGTACGAACT............................................ | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ....................................................................................................TTGAATTTTCCAAATATTCAT............................................................................ | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 |