chrV:3304831-3304888 -
score=-16.30
score=-16.30
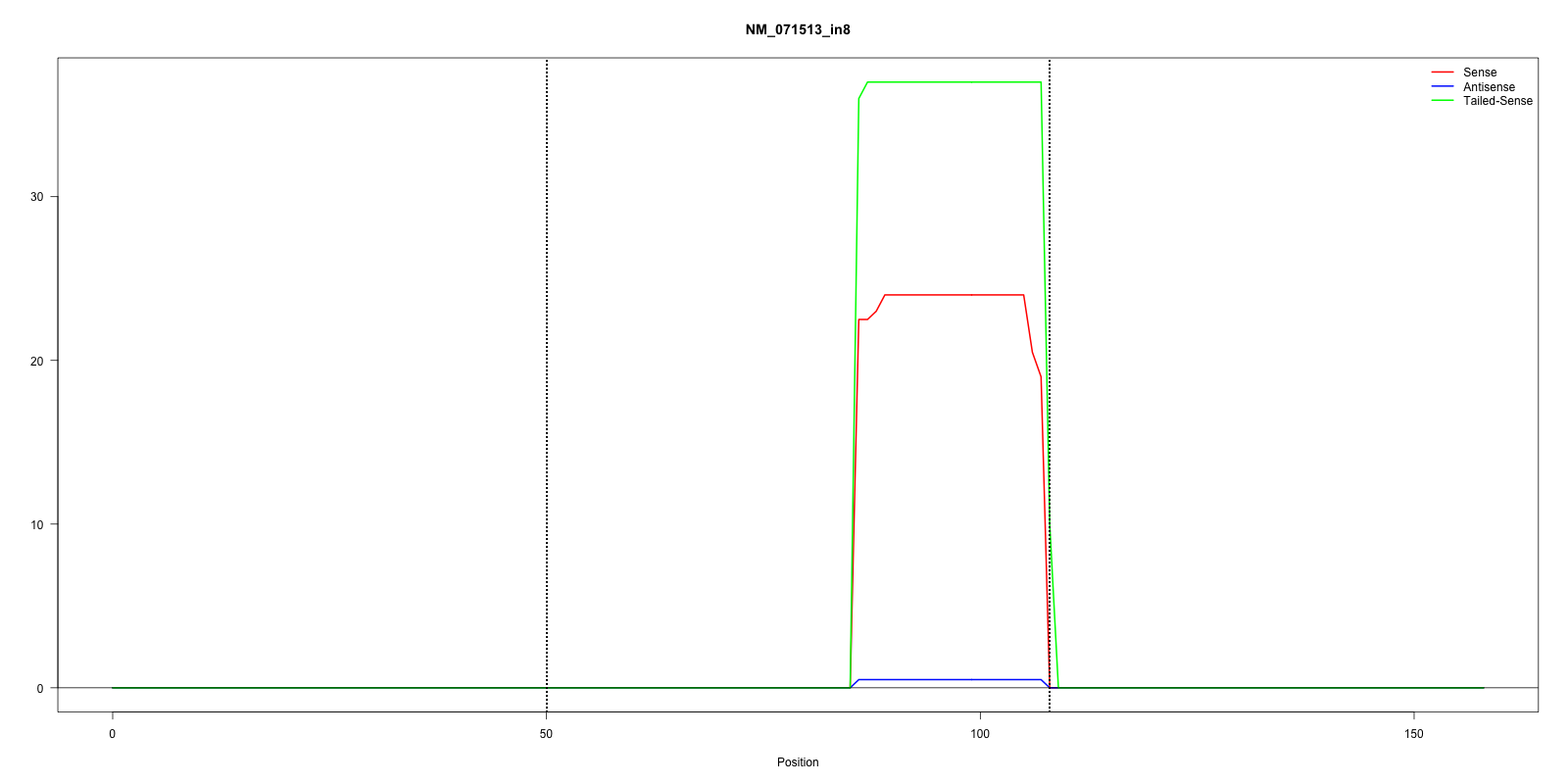
| ID: NM_071513_in8 |
 chrV:3304831-3304888 - score=-16.30 |
 |
| TCTGCCAAAGATAGACGATATGGGCAGTTTTAATGCTAGCGGTCATTGGAGTGAGTAGGACAATTCATATTGAAAATATTGTCTTTTATATAACTGTTCTATTCACAGAACTACTTCGTCGTTATGAAGAATTTGATACACAAAGGCAGCCCATCCAA ..................................................((((((((((((.((.(((.(((((........)))))))))).)))))))))))).................................................... ..................................................51.......................................................108................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM444004 | GSM444001 | GSM444005 | GSM632211 | GSM632210 | GSM509582 | GSM518371 | GSM455402 | GSM444000 | GSM454002 | GSM464611 | GSM632213 | GSM455390 | GSM455389 | GSM642429 | GSM443998 | GSM518370 | GSM336086 | GSM297751 | GSM463029 | GSM632212 | GSM464612 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TATATAACTGTTCTATTCACAC.................................................. | 22 | C | 26.00 | 1.50 | 5.00 | 2.00 | 3.00 | 4.00 | 3.00 | 3.00 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCACAG.................................................. | 22 | 2 | 17.50 | 17.50 | 4.50 | 4.50 | 1.00 | 0.00 | 0.50 | 1.00 | 0.50 | 1.00 | 1.50 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.50 | 0.00 | 0.50 | 0.50 |
| ......................................................................................TATATAACTGTTCTATTCACAGT................................................. | 23 | T | 4.00 | 17.50 | 0.00 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCAC.................................................... | 20 | 2 | 3.50 | 3.50 | 2.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCACACT................................................. | 23 | CT | 3.00 | 1.50 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCACA................................................... | 21 | 2 | 1.50 | 1.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| .........................................................................................ATAACTGTTCTATTCACAG.................................................. | 19 | 2 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCACACC................................................. | 23 | CC | 1.00 | 1.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| .......................................................................................ATATAACTGTTCTATTCACACC................................................. | 22 | CC | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCACATT................................................. | 23 | TT | 1.00 | 1.50 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TATATAACTGTTCTATTCAAAC.................................................. | 22 | AAC | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ........................................................................................TATAACTGTTCTATTCACAG.................................................. | 20 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TCTGCCAAAGATAGACGATATGGGCAGTTTTAATGCTAGCGGTCATTGGAGTGAGTAGGACAATTCATATTGAAAATATTGTCTTTTATATAACTGTTCTATTCACAGAACTACTTCGTCGTTATGAAGAATTTGATACACAAAGGCAGCCCATCCAA ..................................................((((((((((((.((.(((.(((((........)))))))))).)))))))))))).................................................... ..................................................51.......................................................108................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM444004 | GSM444001 | GSM444005 | GSM632211 | GSM632210 | GSM509582 | GSM518371 | GSM455402 | GSM444000 | GSM454002 | GSM464611 | GSM632213 | GSM455390 | GSM455389 | GSM642429 | GSM443998 | GSM518370 | GSM336086 | GSM297751 | GSM463029 | GSM632212 | GSM464612 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TATATAACTGTTCTATTCACAG.................................................. | 22 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |