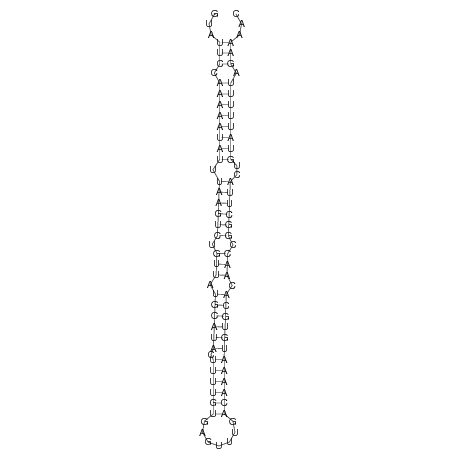
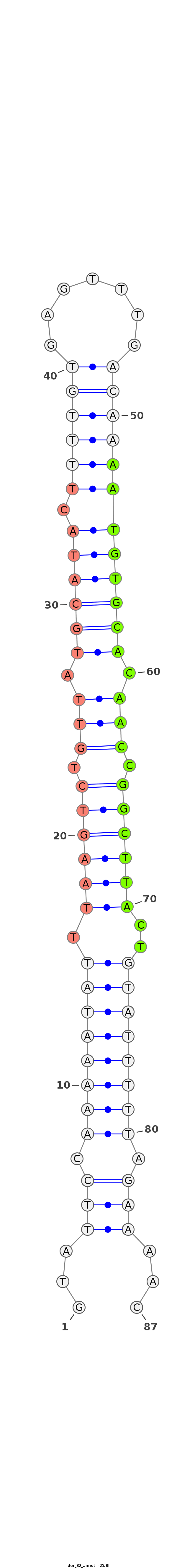
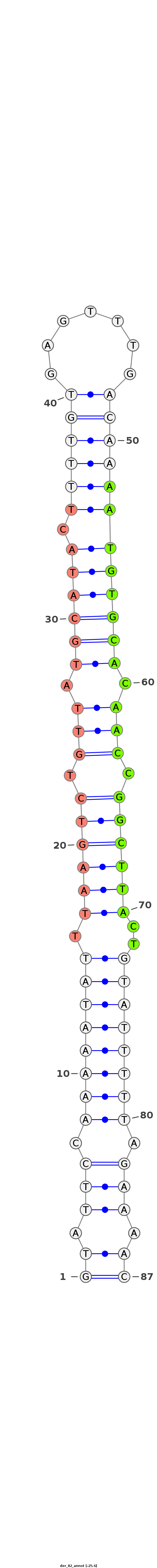
ID:der_82 |
Coordinate:scaffold_4845:10617479-10617536 - |
Confidence:novel |
Type:intergenic |
[View on UCSC Genome Browser] |
| Legend: | mature | star | mismatch in read |
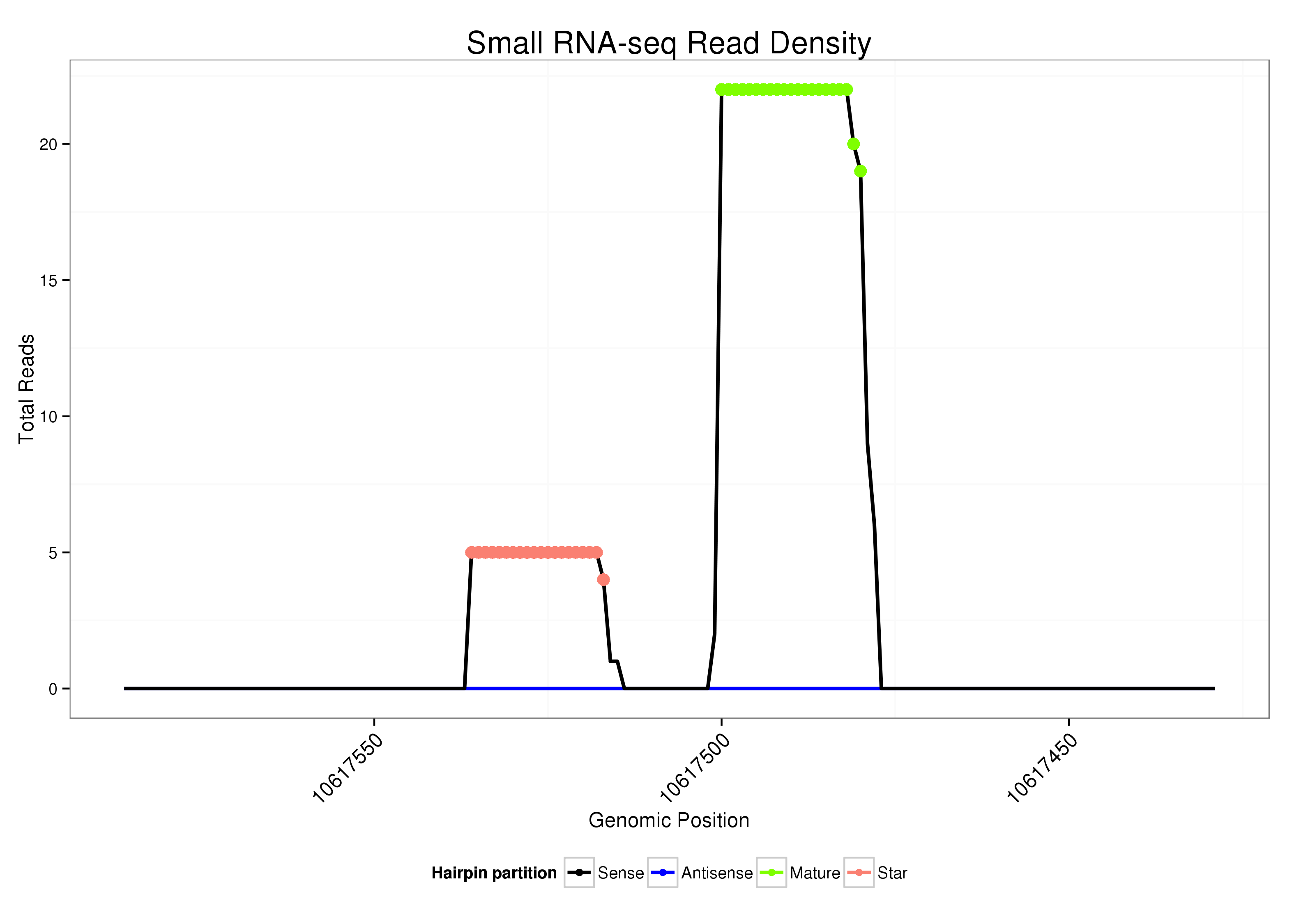
 |
 |
 |
| -25.6 |
 |
intergenic
No Repeatable elements found
|
TTGATTCTGCTTCTTAAATCGCCACCCAACTTATAGTATTCCAAAAATATTTAAGTCTGTTATGCATACTTTTGTGAGTTTGACAAAATGTGCACAACCGGCTTACTGTATTTTTAGAAAACAAACACCTTAGTTGTAAAGAATAAACTAATTGTAAA
***********************************...(((.((((((((.((((((.(((.((((((.((((((.......)))))))))))).))).))))))..)))))))).)))...************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V107 male body malebody |
|---|---|---|---|---|---|---|
| ......................................................................................AATGTGCACAACCGGCTTACT................................................... | 21 | 0 | 1 | 10.00 | 10 | 10 |
| ......................................................................................AATGTGCACAACCGGCTTACTGT................................................. | 23 | 0 | 1 | 6.00 | 6 | 6 |
| ......................................................................................AATGTGCACAACCGGCTTACTG.................................................. | 22 | 0 | 1 | 3.00 | 3 | 3 |
| ..................................................TTAAGTCTGTTATGCATACT........................................................................................ | 20 | 0 | 1 | 3.00 | 3 | 3 |
| .....................................................................................AAATGTGCACAACCGGCTTA..................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 |
| ..................................................TTAAGTCTGTTATGCATAC......................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................AATGTGCACAACCGGCTTAC.................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................TTAAGTCTGTTATGCATACTTT...................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................AATGTGCACAACCGGCTTACTGAA................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................AATGTGCACAACCGGCTTACTGTTAA.............................................. | 26 | 3 | 1 | 1.00 | 1 | 1 |
| .....................................................................................AAATGTGCACAACCGGCTTAA.................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 |
| ..................................................TTAAGTCTGTTATGCATAAT........................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 1 |
| ..................................................TTAAGTCTGTTATGCATAAA........................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 1 |
| .......................................................................................................TACTGTATTTGTAGAAAAA.................................... | 19 | 2 | 10 | 0.10 | 1 | 1 |
| ..................................................TTAAGTCTGTTATGCAGTTT........................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 |
|
TTTACAATTAGTTTATTCTTTACAACTAAGGTGTTTGTTTTCTAAAAATACAGTAAGCCGGTTGTGCACATTTTGTCAAACTCACAAAAGTATGCATAACAGACTTAAATATTTTTGGAATACTATAAGTTGGGTGGCGATTTAAGAAGCAGAATCAA
************************************...(((.((((((((..((((((.(((.((((((((((((.......)))))).)))))).))).)))))).)))))))).)))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 01/18/2014 at 01:38 PM