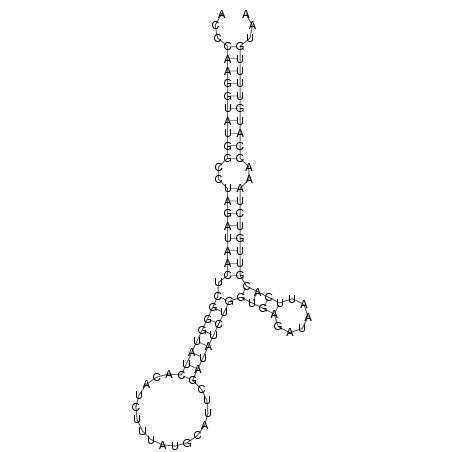
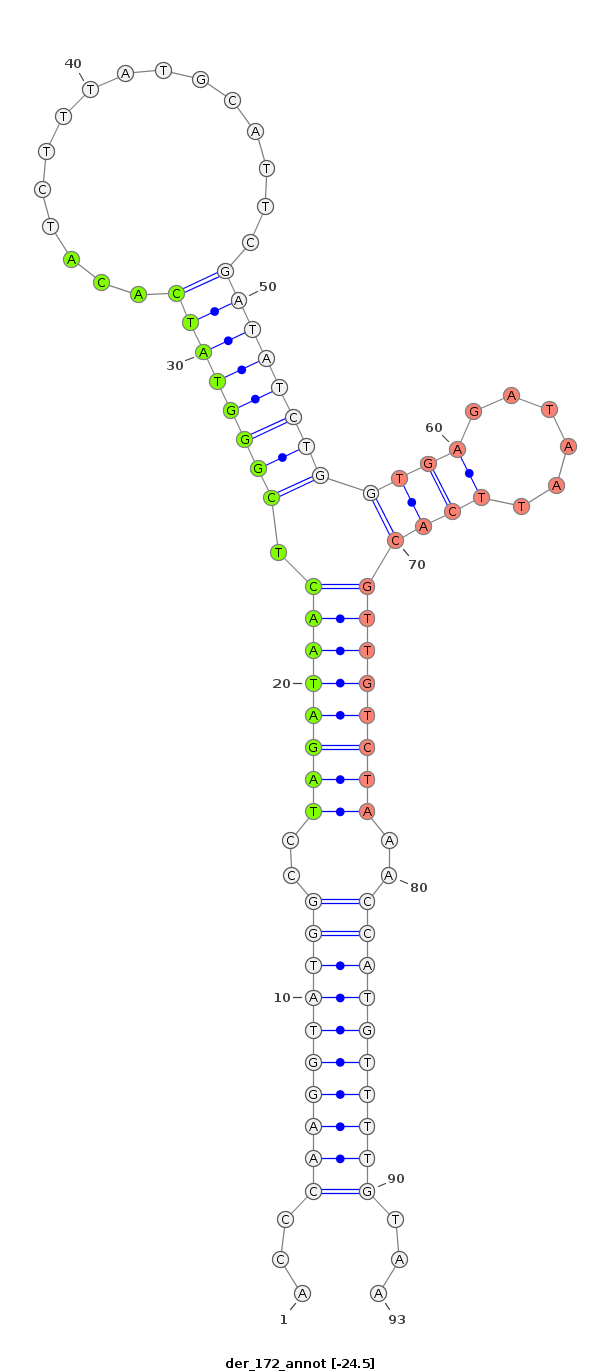
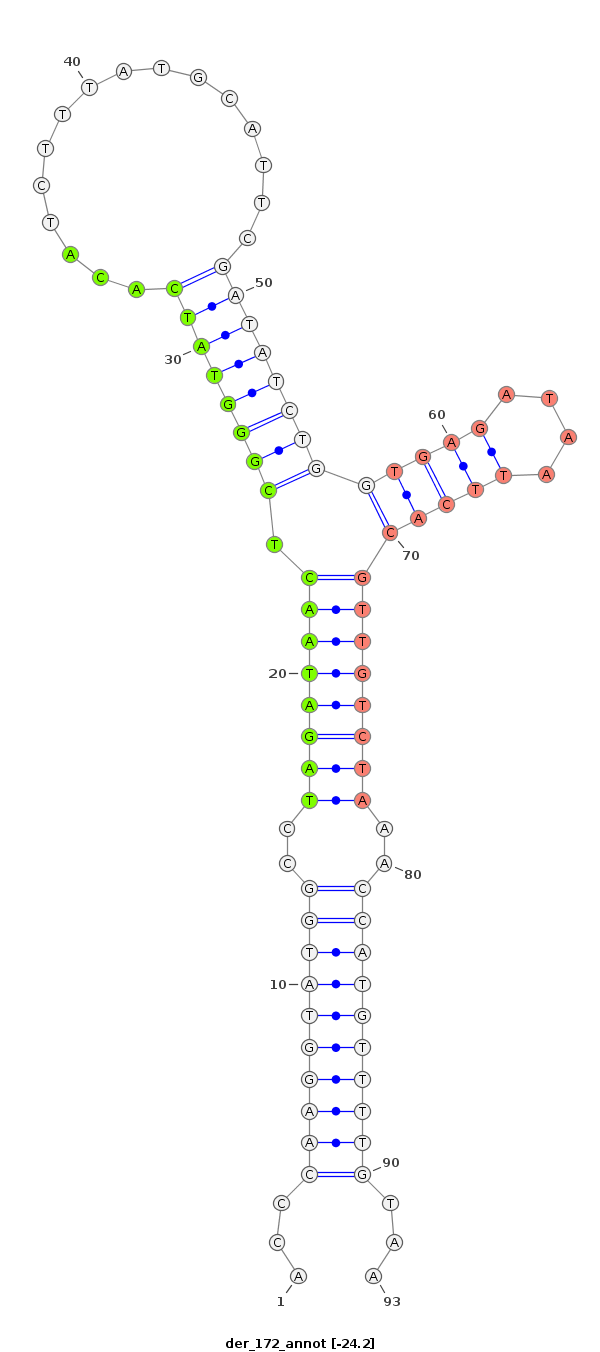
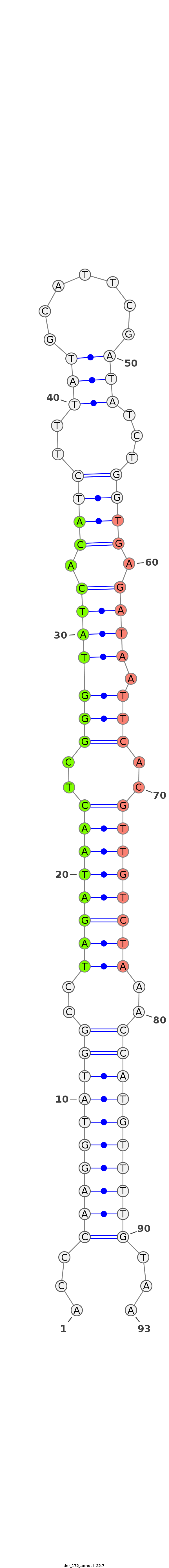
ID:der_172 |
Coordinate:scaffold_4690:9689075-9689177 + |
Confidence:novel |
Type:intergenic |
[View on UCSC Genome Browser] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
| -24.2 | -22.7 |
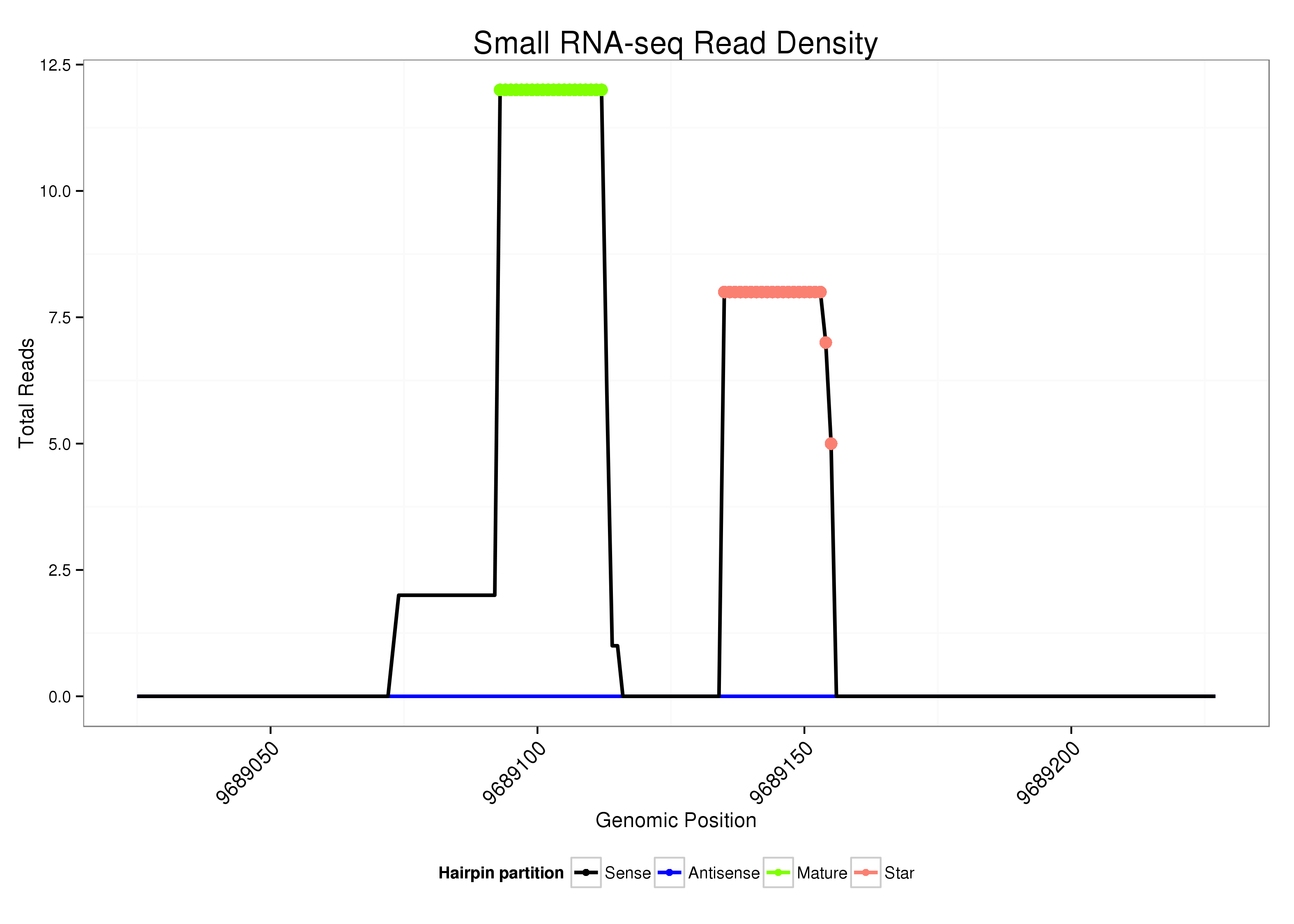
 |
 |
intergenic
No Repeatable elements found
|
GGATCCCGGATTCGGAGTCAACAAACTCTTGTCAAACAAACGAGCAGAATTCAACCCAAGGTATGGCCTAGATAACTCGGGTATCACATCTTTATGCATTCGATATCTGGTGAGATAATTCACGTTGTCTAAACCATGTTTTGTAAGAAATAACGCCTTAACTCGGTCTGCAGCTCAAATAAACTTGGTGTTCAGTTTAAACT
*****************************************************...((((((((((..((((((((.((((((((................))))))))((((......))))))))))))..))))))))))...********************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V107 male body malebody |
|---|---|---|---|---|---|---|
| ....................................................................TAGATAACTCGGGTATCACA................................................................................................................... | 20 | 0 | 1 | 6.00 | 6 | 6 |
| ....................................................................TAGATAACTCGGGTATCACAT.................................................................................................................. | 21 | 0 | 1 | 5.00 | 5 | 5 |
| ..............................................................................................................TGAGATAATTCACGTTGTCTA........................................................................ | 21 | 0 | 1 | 5.00 | 5 | 5 |
| ..............................................................................................................TGAGATAATTCACGTTGTCT......................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 |
| .................................................TTCAACCCAAGGTATGGCC....................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................TAGATAACTAGGGTATCACAT.................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 |
| ..............................................................................................................TGAGATAATTCACGTTGTC.......................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
| ................................................ATTCAACCCAAGGTATGGCC....................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................TAGATAACTCGGGTATCACATCT................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 |
|
AGTTTAAACTGAACACCAAGTTTATTTGAGCTGCAGACCGAGTTAAGGCGTTATTTCTTACAAAACATGGTTTAGACAACGTGAATTATCTCACCAGATATCGAATGCATAAAGATGTGATACCCGAGTTATCTAGGCCATACCTTGGGTTGAATTCTGCTCGTTTGTTTGACAAGAGTTTGTTGACTCCGAATCCGGGATCC
*********************************************************...((((((((((..((((((((((((......))))((((((((................)))))))).))))))))..))))))))))...***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 01/18/2014 at 01:41 PM