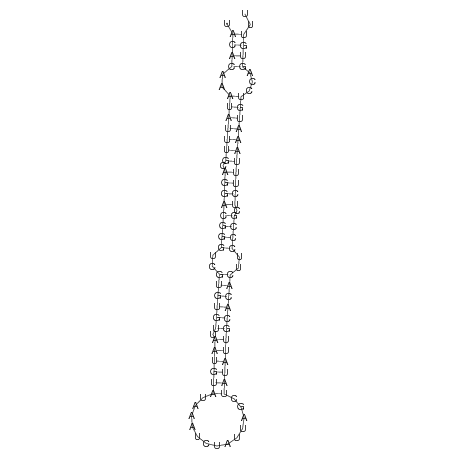
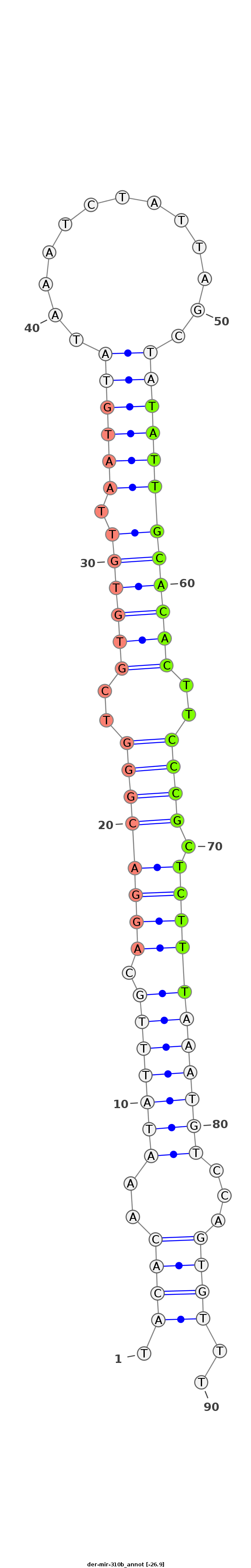
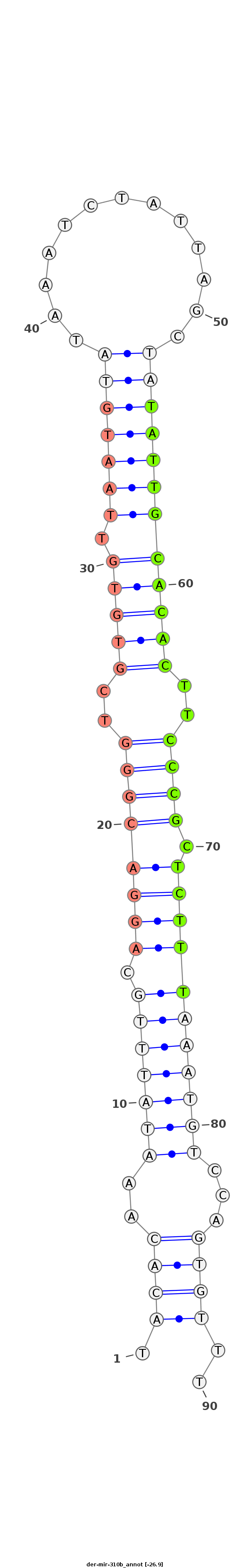
ID:der-mir-310b |
Coordinate:scaffold_4845:10617892-10617979 - |
Confidence:Known |
Type:Unknown |
[View on miRBase] [View on UCSC Genome Browser] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
| -26.9 | -26.4 | -26.4 |
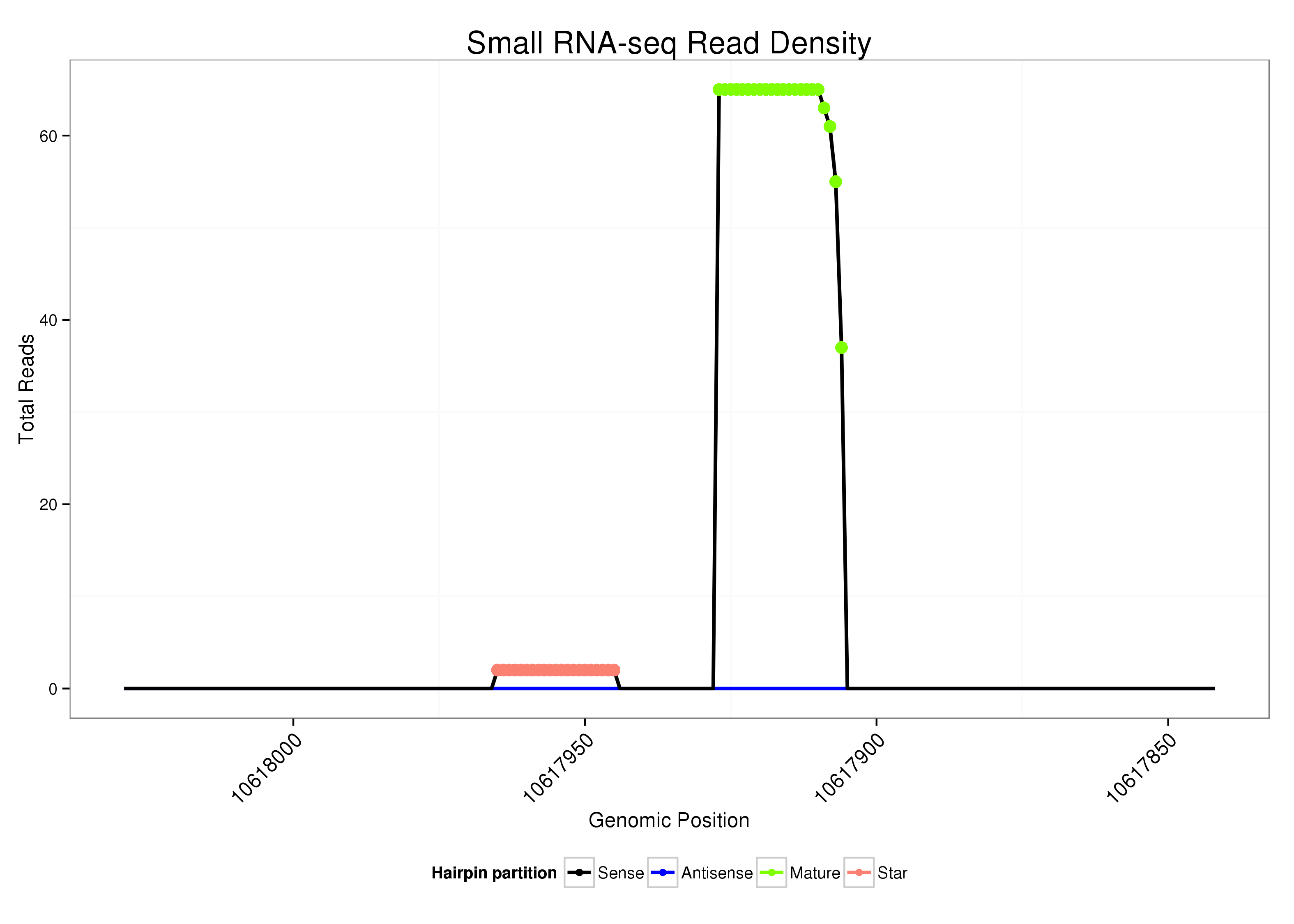
 |
 |
 |
pre_miRNA [Dere\mir-310b-RM]; miRNA [Dere\miR-310b-RA]; pre_miRNA [Dere\mir-311c-RM]; miRNA [Dere\miR-311c-RA]
No Repeatable elements found
|
TTCACCAGCCTGAAAATATCAAGGACTTACATCAAAAAGCGCTTGCAAATACACAAATATTTGCAGGACGGGTCGTGTGTTAATGTATAAATCTATTAGCTATATTGCACACTTCCCGCTCTTTAAATGTCCAGTGTTTCAACTTGAATATCATTTCGAGTTCAATGGAATTCGACAGTTCGTTCTTT
*************************************************.((((..(((((((.((((((((..((((((.((((((.............))))))))))))..)))).)))))))))))...))))..************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V107 male body malebody |
|---|---|---|---|---|---|---|
| ......................................................................................................TATTGCACACTTCCCGCTCTTT................................................................ | 22 | 0 | 1 | 37.00 | 37 | 37 |
| ......................................................................................................TATTGCACACTTCCCGCTCTT................................................................. | 21 | 0 | 1 | 18.00 | 18 | 18 |
| ......................................................................................................TATTGCACACTTCCCGCTCT.................................................................. | 20 | 0 | 1 | 6.00 | 6 | 6 |
| ......................................................................................................TATTGCACACTTCCCGCTCTTTT............................................................... | 23 | 1 | 1 | 3.00 | 3 | 3 |
| ......................................................................................................TATTGCACACTTCCCGCT.................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 |
| ......................................................................................................TATTGCACACTTCCCGCTC................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 |
| ................................................................AGGACGGGTCGTGTGTTAATG....................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 |
| ......................................................................................................TATTGCACACTTCCCGCTCTTTAT.............................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................TATTGCACACTTCCCGCTCTTTG............................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................TATTGCACACTTCCCGCTCG.................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................TATTGCACACTTCCCGCTTA.................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 |
|
AAAGAACGAACTGTCGAATTCCATTGAACTCGAAATGATATTCAAGTTGAAACACTGGACATTTAAAGAGCGGGAAGTGTGCAATATAGCTAATAGATTTATACATTAACACACGACCCGTCCTGCAAATATTTGTGTATTTGCAAGCGCTTTTTGATGTAAGTCCTTGATATTTTCAGGCTGGTGAA
*************************************************..((((...(((((((((((.((((..((((((((((((.............)))))).))))))..)))))))).)))))))..)))).************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|---|---|---|---|---|
| ..AGAACGAACCGGCGAGTT........................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 |
Generated: 01/18/2014 at 01:55 PM