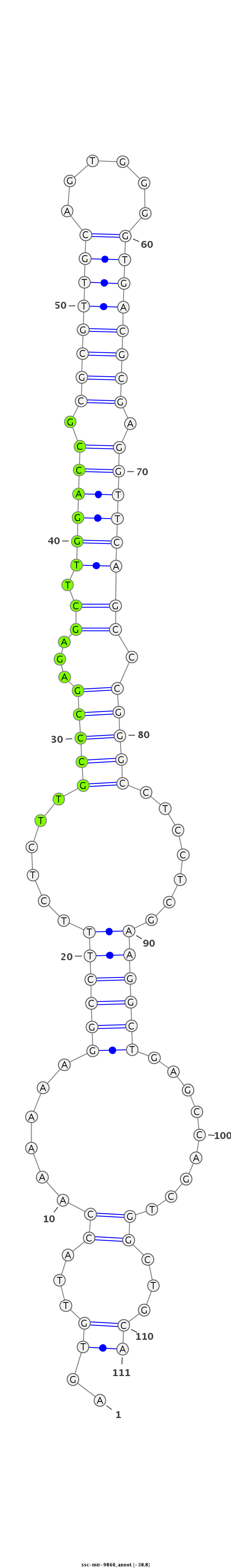
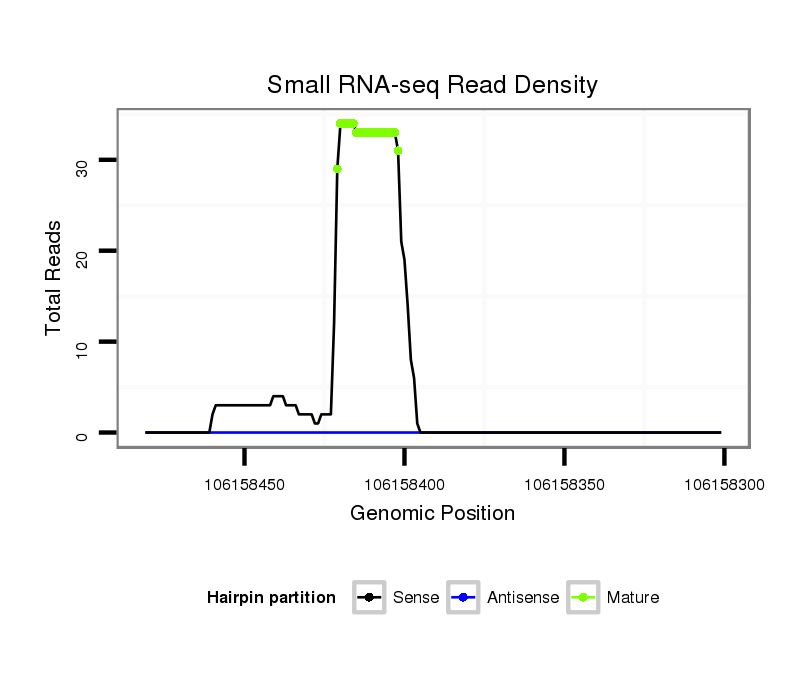
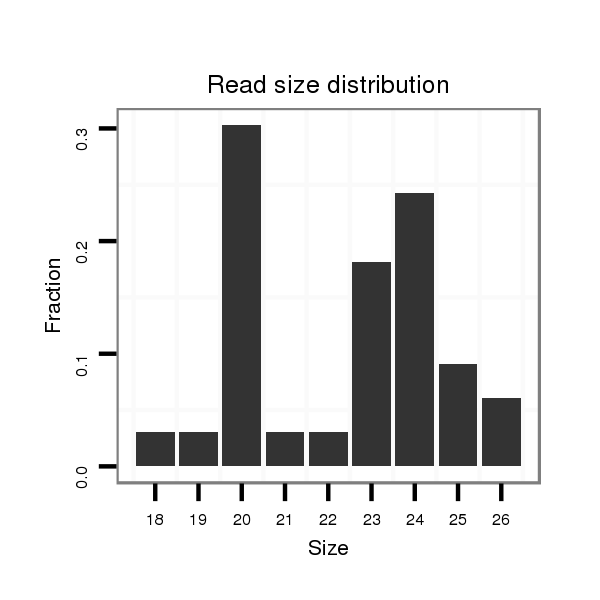
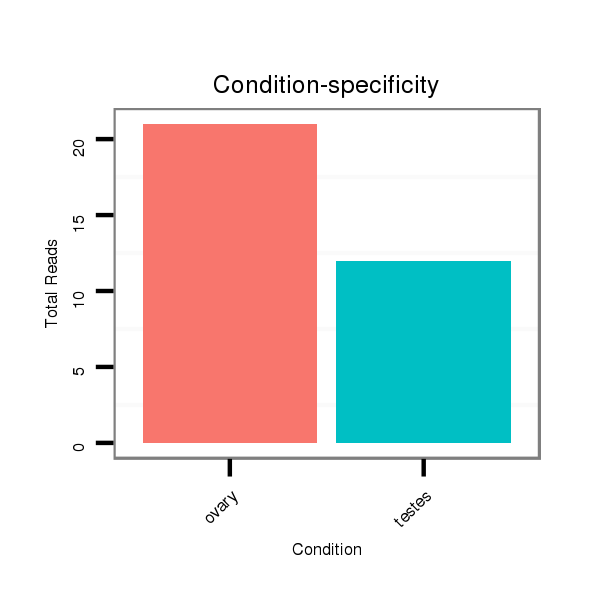
ID:ssc-mir-9860 |
Coordinate:chr7:106158351-106158431 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -46.7 | -46.7 | -46.0 | -46.0 |
 |
 |
 |
 |
|
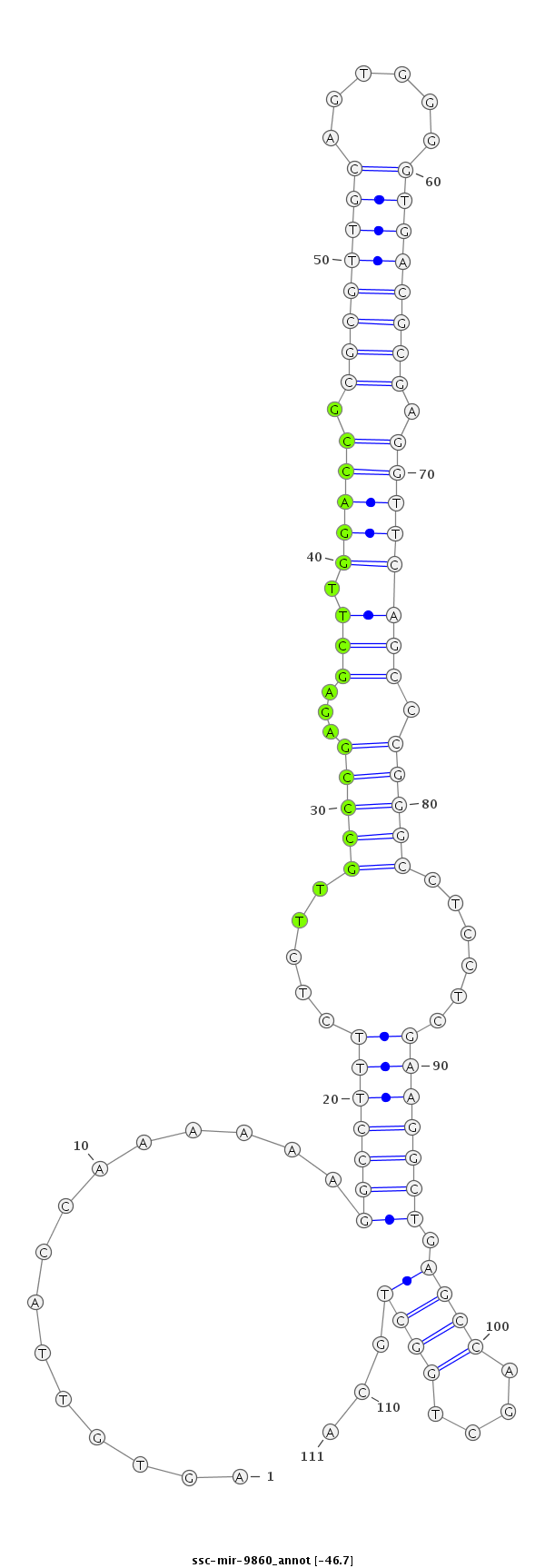
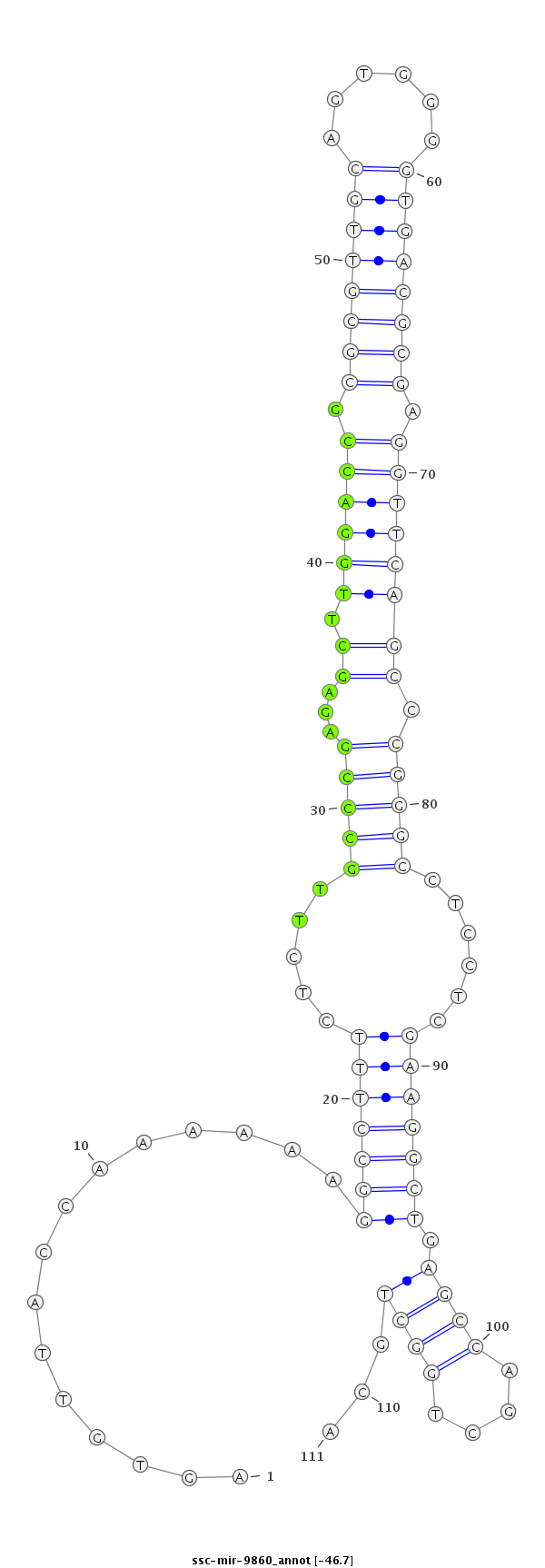
GGCCAAAGAACCGGGTCCTGCTTACTGCTGGCTTGAGTGTTACCAAAAAAGGCCTTTCTCTTGCCCGAGAGCTTGGACCGCGCGTTGCAGTGGGGTGACGCGAGGTTCAGCCCGGGCCTCCTCGAAGGCTGAGCCAGCTGGCTGCAGGATGCCAGGCCAGAGTCCCCCGCCCGCCCCATCT
***********************************..((...((......((((((......(((((...((.((((((.((((((((......)))))))).)))))))).))))).......)))))).........))...))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR1274763 ovary |
SRR139075 testes |
SRR066810 testes |
SRR1274764 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TTGCCCGAGAGCTTGGACCG..................................................................................................... | 20 | 0 | 1 | 9.00 | 9 | 0 | 7 | 1 | 0 | 1 |
| ...........................................................CTTGCCCGAGAGCTTGGACCGCGC.................................................................................................. | 24 | 0 | 1 | 5.00 | 5 | 4 | 0 | 0 | 0 | 1 |
| ...........................................................CTTGCCCGAGAGCTTGGACCGCG................................................................................................... | 23 | 0 | 1 | 4.00 | 4 | 2 | 1 | 0 | 1 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGCGT................................................................................................ | 25 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 0 | 1 |
| .............................................................TGCCCGAGAGCTTGGACCGCGCGT................................................................................................ | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| .............................................................TGCCCGAGAGCTTGGACC...................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................CTTGTCCGAGAGCTTGGACCGCG................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGT.................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................TGCCCGAGAGCTTGGACCGCGCG................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGCGA................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................AAAAAGGTCTTTCTCTTGCCCGAGAGC............................................................................................................. | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCG................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................TTACTGCTGGCTTGAGTGTTACCAAAAAAGGC................................................................................................................................ | 32 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................TACTGCTGGCTTGAGTGTTACC......................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGT.................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................TACCAAAAAAGGCCTTTCTCTTGCCC................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGCGGAT.............................................................................................. | 27 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCACGA................................................................................................ | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGCG................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGCGTT............................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................TGCCCGAGAGCTTGGACCGC.................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................TTACTGCTGGCTTGAGTGTTACCAAAA..................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................TGCCCGAGAGCTTGGACCGCGCA................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACC...................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTGCCCGAGAGCTTGGACCGCGC.................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................CTTGCCCGAGAGCTTGGACCG..................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................TTCTCTTGCCCGAGAGCTTGGACCGC.................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
|
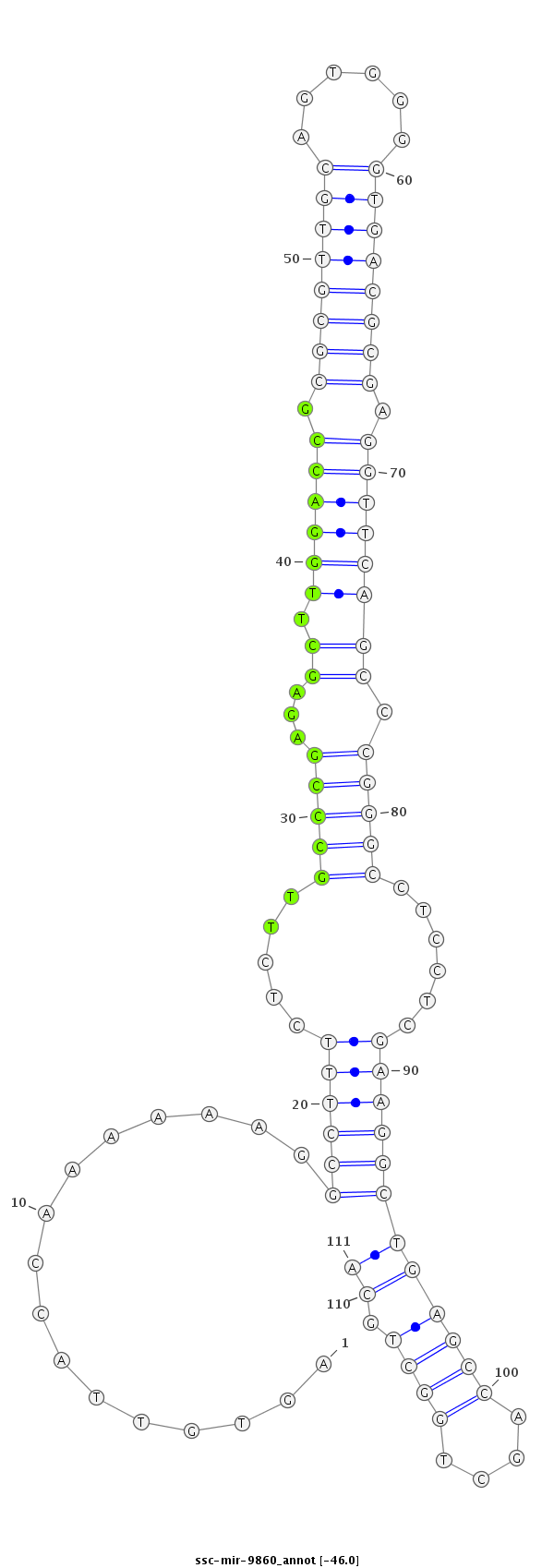
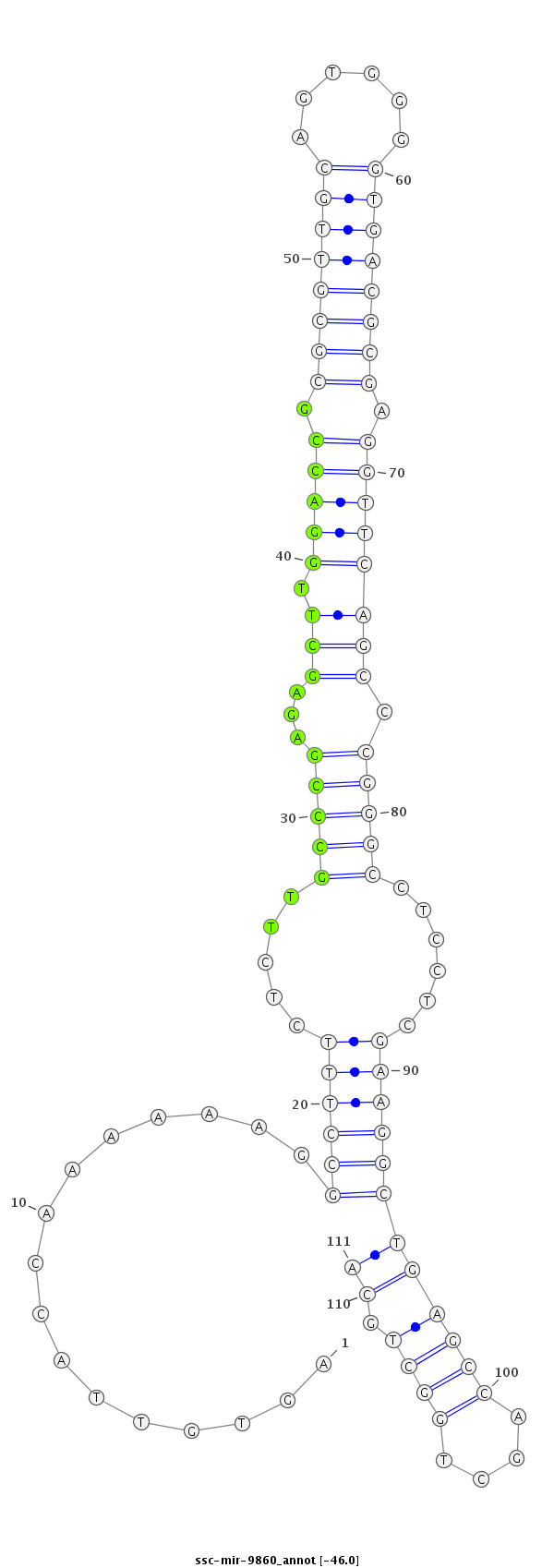
CCGGTTTCTTGGCCCAGGACGAATGACGACCGAACTCACAATGGTTTTTTCCGGAAAGAGAACGGGCTCTCGAACCTGGCGCGCAACGTCACCCCACTGCGCTCCAAGTCGGGCCCGGAGGAGCTTCCGACTCGGTCGACCGACGTCCTACGGTCCGGTCTCAGGGGGCGGGCGGGGTAGA
***********************************..((...((......((((((......(((((...((.((((((.((((((((......)))))))).)))))))).))))).......)))))).........))...))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:40 AM