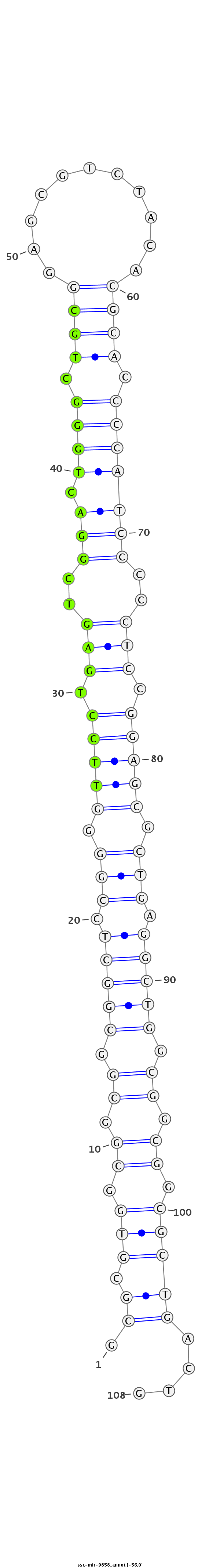
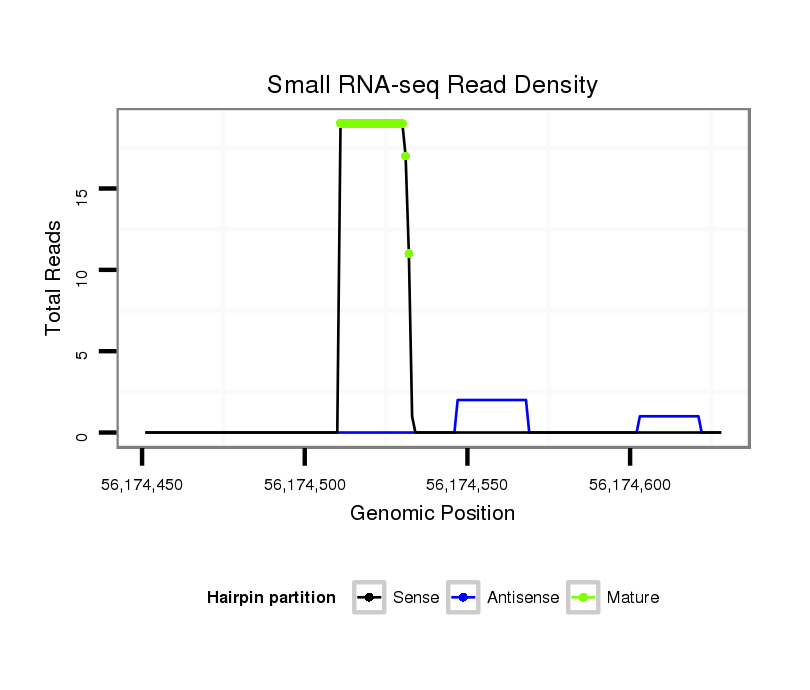
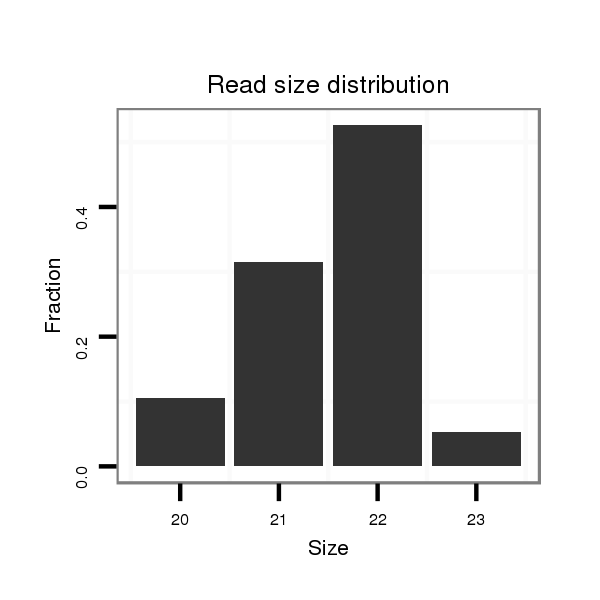
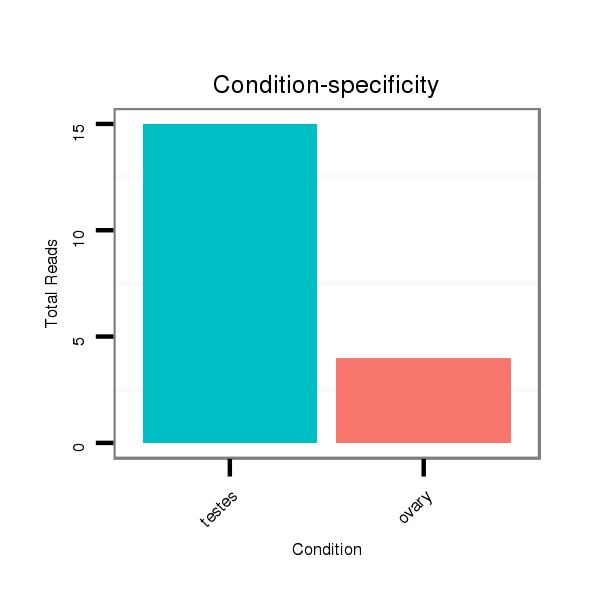
ID:ssc-mir-9858 |
Coordinate:chr4:56174501-56174578 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -55.4 |
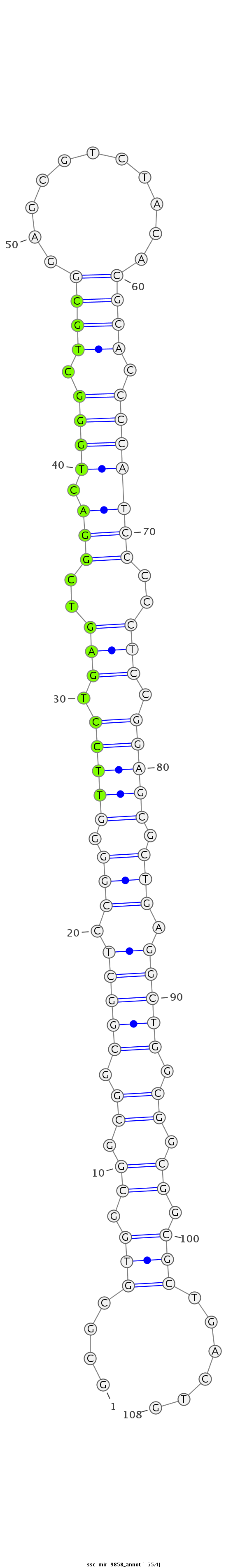 |
|
GTCCCTGGGCCTCCACGAGGCTCCGTGCTCCCCGCGCGCGTGGCGGCGGCGGCTCCGGGGTTCCTGAGTCGGACTGGGCTGCGGAGCGTCTACACGCACCCCATCCCCCTCCGGAGCGCTGAGGCTGGCGGCGGCGCTGACTGCTGAGGACCCGCGTCCGGTATCTCTGATCCCCTGT
***********************************.((.(((.((.((.(((((.(((.(((((.(((..(((.((((.((((...........)))).)))))))..))).))))).))).))))).)).)).)))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR1274763 ovary |
SRR1274764 testes |
SRR139076 testes |
SRR066809 ovary |
SRR066810 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TTCCTGAGTCGGACTGGGCTGC................................................................................................ | 22 | 0 | 1 | 10.00 | 10 | 4 | 2 | 0 | 1 | 1 | 2 |
| ............................................................TTCCTGAGTCGGACTGGGCTG................................................................................................. | 21 | 0 | 1 | 6.00 | 6 | 5 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCGA.............................................................................................. | 24 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTAT................................................................................................ | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCGT.............................................................................................. | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCAA.............................................................................................. | 24 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCT.................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCA............................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGTAT.............................................................................................. | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGAA............................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCGTT............................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCTA.............................................................................................. | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGAAGT............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCAC.............................................................................................. | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCCGACTGGGCTGC................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTT................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCAT.............................................................................................. | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTGCG............................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCTTAT............................................................................................... | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TTCCTGAGTCGGACTGGGCAAT................................................................................................ | 22 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CAGGGACCCGGAGGTGCTCCGAGGCACGAGGGGCGCGCGCACCGCCGCCGCCGAGGCCCCAAGGACTCAGCCTGACCCGACGCCTCGCAGATGTGCGTGGGGTAGGGGGAGGCCTCGCGACTCCGACCGCCGCCGCGACTGACGACTCCTGGGCGCAGGCCATAGAGACTAGGGGACA
***********************************.((.(((.((.((.(((((.(((.(((((.(((..(((.((((.((((...........)))).)))))))..))).))))).))).))))).)).)).)))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139076 testes |
SRR139075 testes |
SRR066810 testes |
SRR066809 ovary |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................GTGGGGTAGGGGGAGGCCTCGC............................................................ | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................GCGCAGGCCATAGAGACTA....... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................CGCACCGCCGCCGCGGA............................................................................................................................ | 17 | 1 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................................................TCGCGCCTCCGGCCGGCGCCGC.......................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 09/01/2015 at 11:35 AM