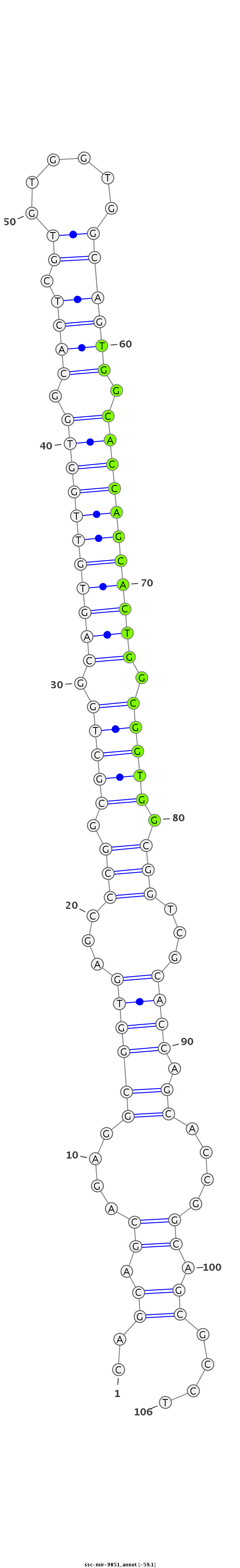
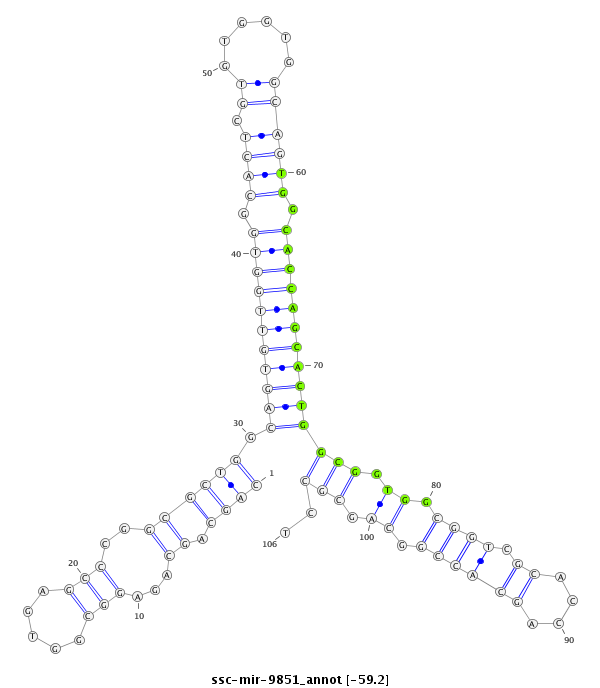
ID:ssc-mir-9851 |
Coordinate:chr14:138325833-138325908 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -59.2 |
 |
|
TGACACAGGCAGTAGCTCCAGCAACGTGATGCCAGCAGCAGCAGAGGCGGTGAGCCCGGCGCTGGCAGTGTTGGTGGCACTCGTGTGGTGGCAGTGGCACCAGCACTGGCGGTGGCGGTCGCACCAGCACCGGCAGCGCCTGTACCAGCAGGAGATGCCACTGCACCTTTGAAGTG
***********************************..((.((....((((((...(((.(((((.(((((((((((.((((.((......)))))).))))))))))).))))).)))...)))).))....)).))....*********************************** |
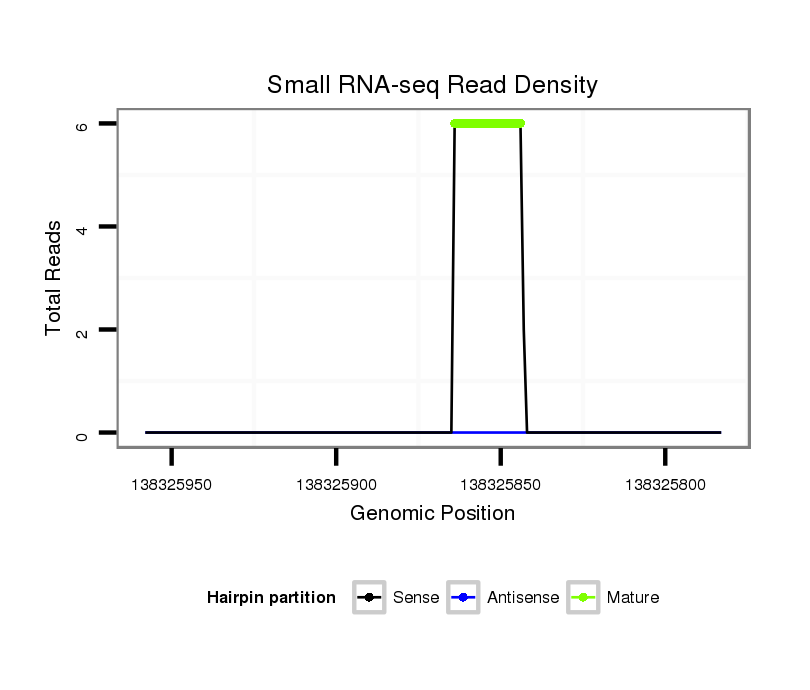
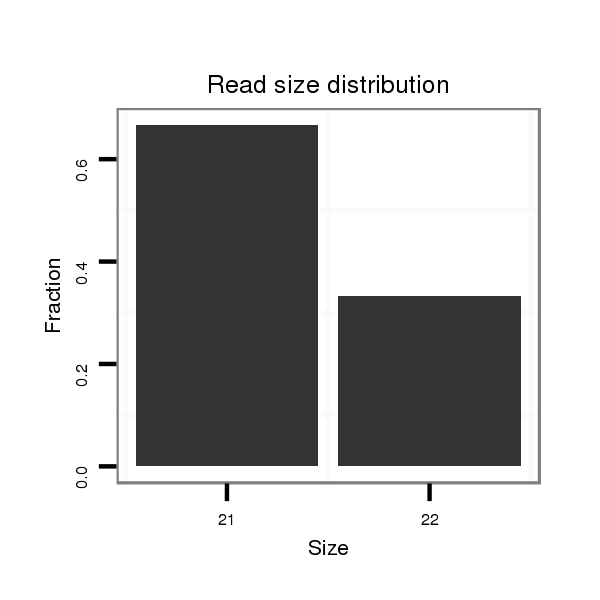
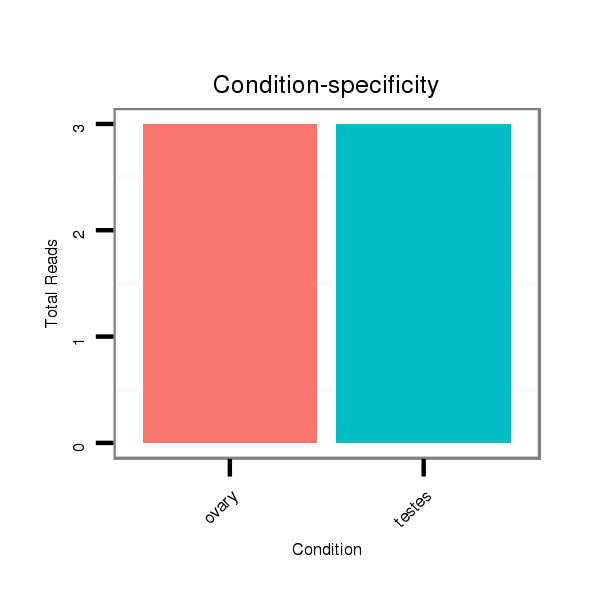
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1274763 ovary |
SRR139075 testes |
SRR1274764 testes |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................TGGCACCAGCACTGGCGGTGG............................................................. | 21 | 0 | 1 | 4.00 | 4 | 2 | 0 | 2 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGC............................................................ | 22 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGCT........................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGAT............................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................TGGCACCGGCACTGGCGGTGG............................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGA............................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGAT........................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGT............................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGCA........................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGTGGCTT.......................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGGCACCAGCACTGGCGGAA.............................................................. | 20 | 2 | 6 | 0.33 | 2 | 0 | 1 | 0 | 1 |
| ....................................CGCAGCAGAGGCGGTGA........................................................................................................................... | 17 | 1 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
ACTGTGTCCGTCATCGAGGTCGTTGCACTACGGTCGTCGTCGTCTCCGCCACTCGGGCCGCGACCGTCACAACCACCGTGAGCACACCACCGTCACCGTGGTCGTGACCGCCACCGCCAGCGTGGTCGTGGCCGTCGCGGACATGGTCGTCCTCTACGGTGACGTGGAAACTTCAC
***********************************..((.((....((((((...(((.(((((.(((((((((((.((((.((......)))))).))))))))))).))))).)))...)))).))....)).))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:27 AM