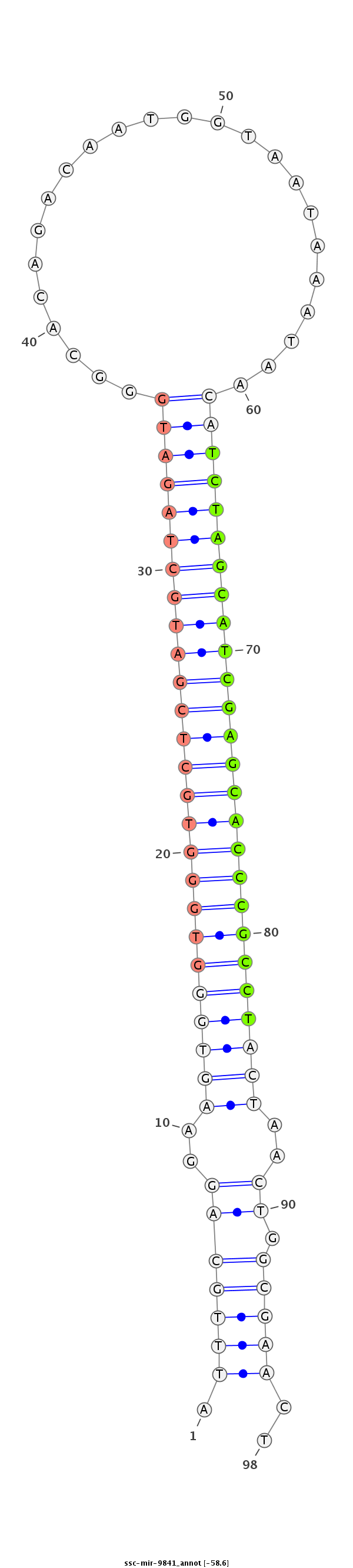
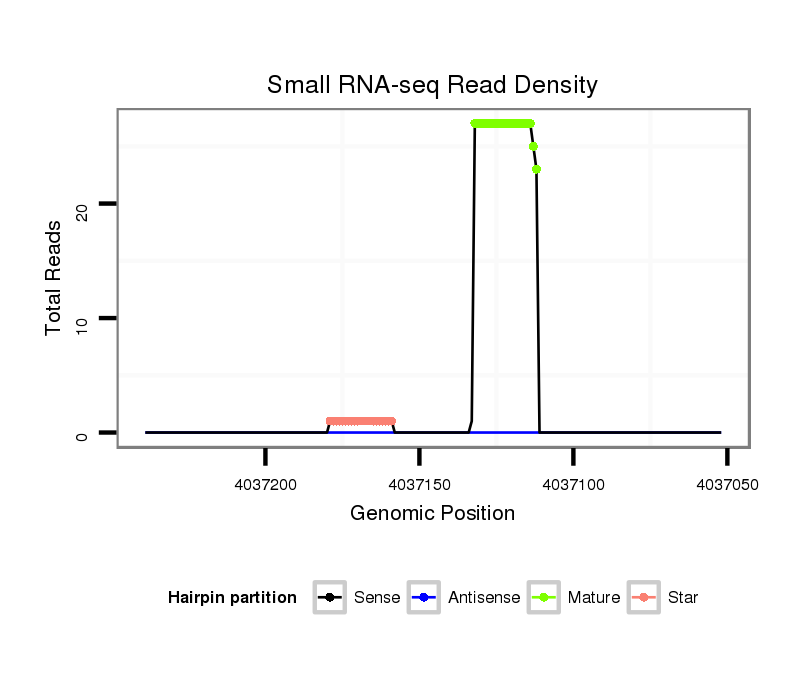
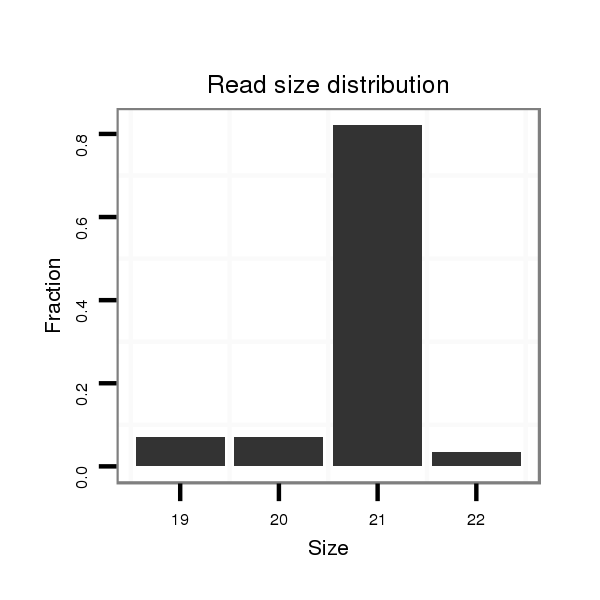
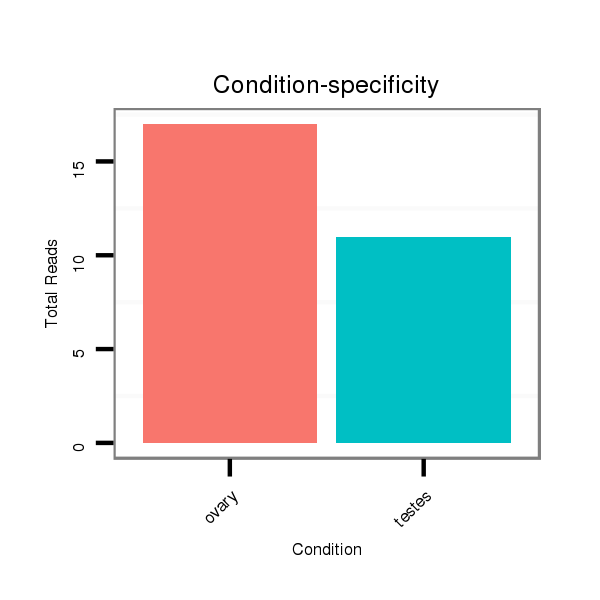
ID:ssc-mir-9841 |
Coordinate:chr7:4037102-4037189 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
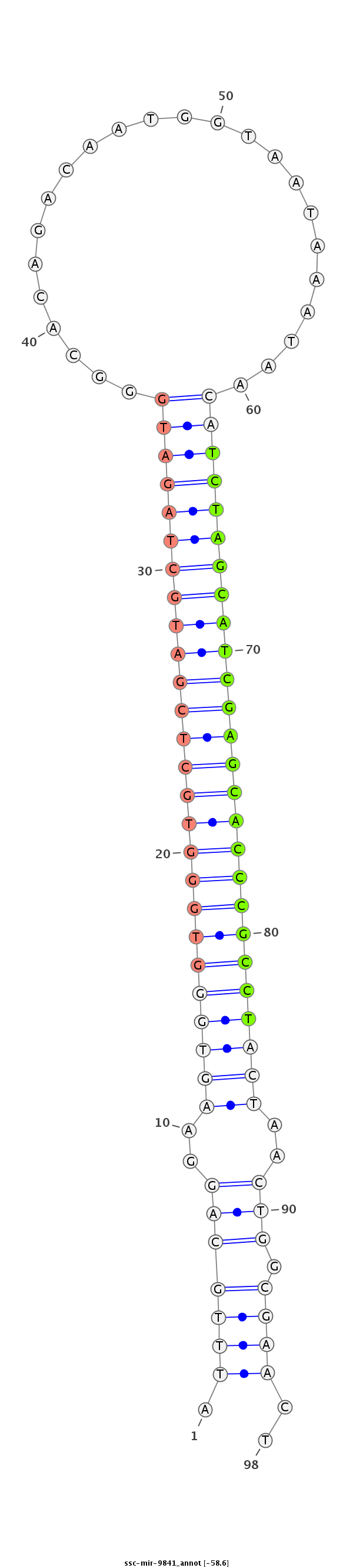
| -58.6 | -58.4 | -58.4 |
 |
 |
 |
|
CTGCAGTCACAGGGAAGCCCAGCACGGAGGATTGATTGGCAAGGAATTTGCAGGAAGTGGGTGGGTGCTCGATGCTAGATGGGCACAGACAATGGTAATAAATAACATCTAGCATCGAGCACCCGCCTACTAACTGGCGAACTTTCTAACGATGATGACAGTAGTGTGGCTTTACAGTCCCAGGGAAA
*********************************************.(((((((..((((((((((((((((((((((((((........................))))))))))))))))))))))))))..)).)))))..********************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR139075 testes |
SRR1274763 ovary |
SRR1274764 testes |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TCTAGCATCGAGCACCCGCCT............................................................ | 21 | 0 | 1 | 22.00 | 22 | 13 | 6 | 1 | 0 | 2 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTAT.......................................................... | 23 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTTT.......................................................... | 23 | 2 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTT........................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCATT.......................................................... | 23 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGC.............................................................. | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCC............................................................. | 20 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTTA.......................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCTTAT.......................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GTAGGTGCTCGATGCTAGATGGGCA....................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................GTGGGTGCTCGATGCTAGATG........................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGTCA............................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GTGGGAGCTCGATGCTAGATG........................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCAAA.......................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................GTGGGTGCTCGATGCTAGATGA.......................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCAA........................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCAGCC......................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................ATCTAGCATCGAGCACCCGCCTAT.......................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTAA.......................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCAAT........................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................GTAGGTGCTCGATGCTAGATG........................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTATT......................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCGTCGAGCACCCGCCTTT.......................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCCTTTT......................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CTAGCATCGAGCACCCGCCTTT.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ATCTAGCATCGAGCACCCGCCT............................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGCAAA........................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCTAGCATCGAGCACCCGT.............................................................. | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................ATCTAGCATCGAGCACCCTCC............................................................. | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
|
GACGTCAGTGTCCCTTCGGGTCGTGCCTCCTAACTAACCGTTCCTTAAACGTCCTTCACCCACCCACGAGCTACGATCTACCCGTGTCTGTTACCATTATTTATTGTAGATCGTAGCTCGTGGGCGGATGATTGACCGCTTGAAAGATTGCTACTACTGTCATCACACCGAAATGTCAGGGTCCCTTT
*********************************************.(((((((..((((((((((((((((((((((((((........................))))))))))))))))))))))))))..)).)))))..********************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
|---|---|---|---|---|---|---|
| ...........................................................CCTCCCACGAGCTACGATCTA............................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 1 |
Generated: 09/01/2015 at 11:39 AM