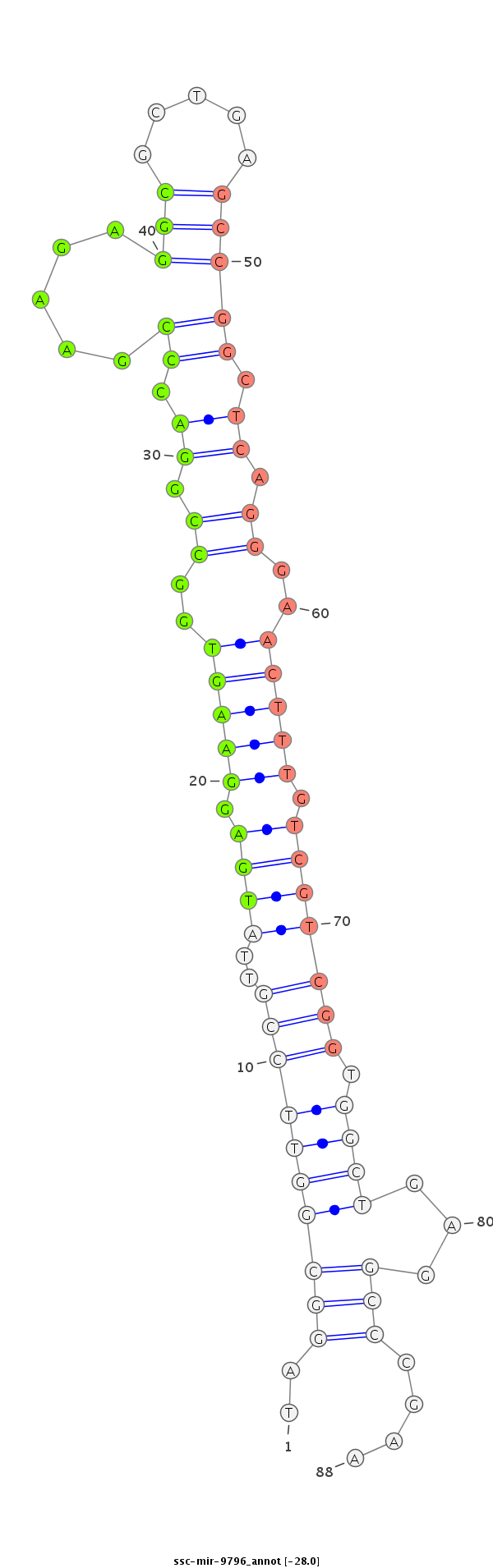


ID:ssc-mir-9796 |
Coordinate:chr3:6616431-6616522 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -27.9 | -27.8 | -27.8 |
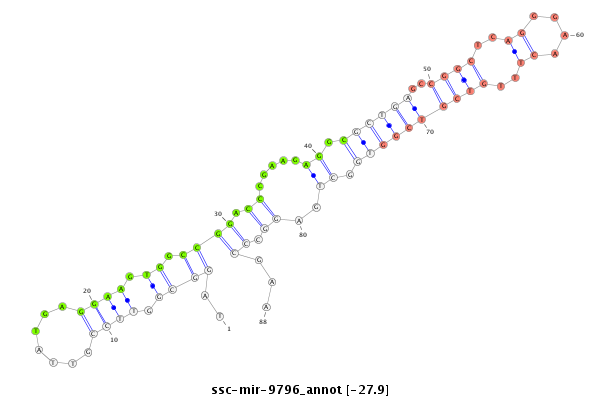 |
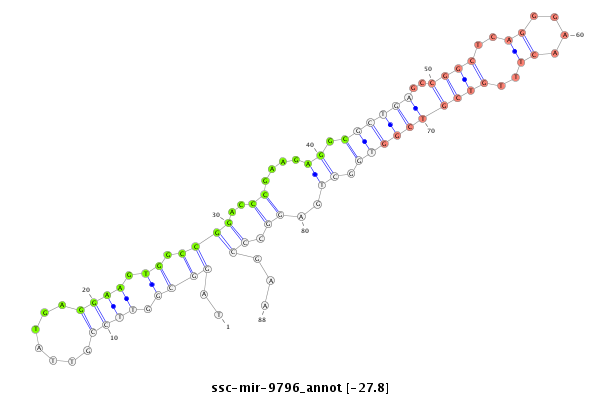 |
 |
|
GTCGGGAGTCCAACCCAACCGGGGACTGTCAGGATCCCGAAATTTCTCGAGGGCATTTTAGGCGGTTCCGTTATGAGGAAGTGGCCGGACCCGAAGAGGCGCTGAGCCGGCTCAGGGAACTTTGTCGTCGGTGGCTGAGGCCCGAAATGCACTCGAAAGAGCAGATCCTAGAGCTGCTGGTGCTGGAGCAGT
**********************************************************..((((((((((..((((.(((((..((.((.((.....(((.....))))).)).))..))))).))))))).))))...)))....********************************************** |
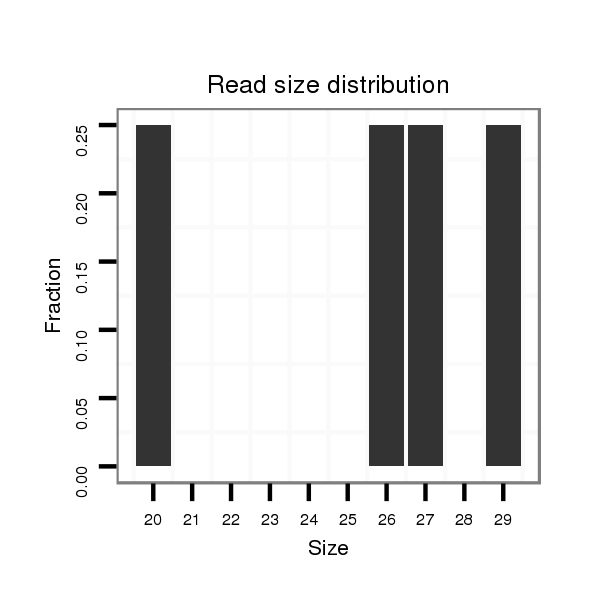
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139076 testes |
SRR1274764 testes |
SRR066810 testes |
SRR139075 testes |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................ACTTTGTCGTCGGTGGCTGAGGCCCGAAA............................................. | 29 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ......................................GAAATTTCTCGAGGGCATTTTAGGCGG............................................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .......................................AAATTTCTCGAGGGCATTTTAGGCGG............................................................................................................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .........................................................................TGAGGAAGTGGCCGGACCCGAAGAGGC............................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................AAATTTCTCGAGGGCATTTTAGGCGGT.............................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................GCCGGACCCGAAGAGGCGCT......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................................................................ACTTTGTCGTCGGTGGCTGAGGCCCGAAATT........................................... | 31 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................TCGGTGGCTGAGGCCCGAAATGCACTCGAA................................... | 30 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................AATTTCTCGAGGGCATTTTAGGCGGTT............................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................AACTTTGTCGTCGGTGGCTGAGGCCCGAAA............................................. | 30 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................................ACTTTGTCGTCGGTGGCTGAGGCCCGAAATGCAC........................................ | 34 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................TGAGGAAGTGGCCGGACCCGAAGAGGCGC.......................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................GCCGGCTCAGGGAACTTTGTCGTCGG............................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...........AACCCAACCGGGGACTGTCAGG............................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................TGTCGTCGGTGGCTGAGGCCCGAA.............................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .......................................AAATTTCTCGAGGGCATTTTAGGCGGTT............................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
|
CAGCCCTCAGGTTGGGTTGGCCCCTGACAGTCCTAGGGCTTTAAAGAGCTCCCGTAAAATCCGCCAAGGCAATACTCCTTCACCGGCCTGGGCTTCTCCGCGACTCGGCCGAGTCCCTTGAAACAGCAGCCACCGACTCCGGGCTTTACGTGAGCTTTCTCGTCTAGGATCTCGACGACCACGACCTCGTCA
**********************************************..((((((((((..((((.(((((..((.((.((.....(((.....))))).)).))..))))).))))))).))))...)))....********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
|---|---|---|---|---|---|---|
| ..........................................................................................................GGCCGAGTCCCTTGAAACAGC................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:33 AM