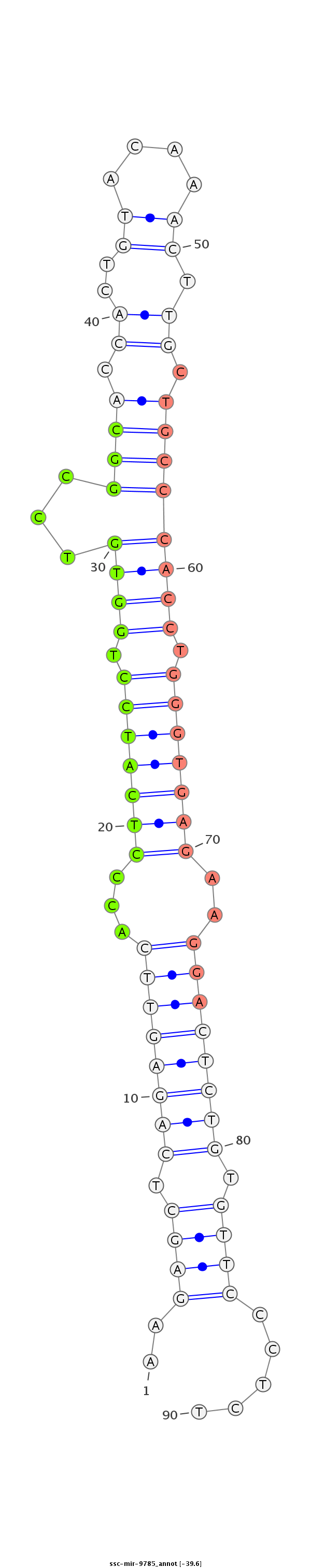
ID:ssc-mir-9785 |
Coordinate:chr3:22233412-22233489 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
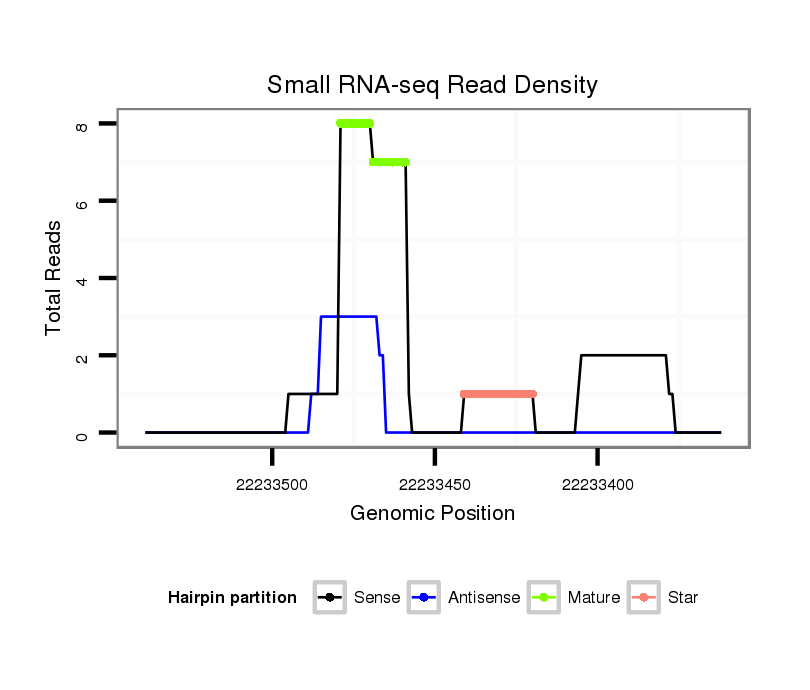
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -39.5 | -38.6 |
 |
 |
|
CATTTGGCCACCCTAGGTAGCACACACCCAGGCGATCTTGCAAGTAAGAGCTCAGAGTTCACCCTCATCCTGGTGTCCGGCACCACTGTACAAACTTGCTGCCCACCTGGGTGAGAAGGACTCTGTGTTCCCTCTGTGTTTCTCTCCATGAAATAAGTCCCTCATGTTGTAAGACTCT
*********************************************..((((.((((((((...(((((((.((((...((((.((..((....)).)).)))))))).)))))))..)))))))).)))).....******************************************* |
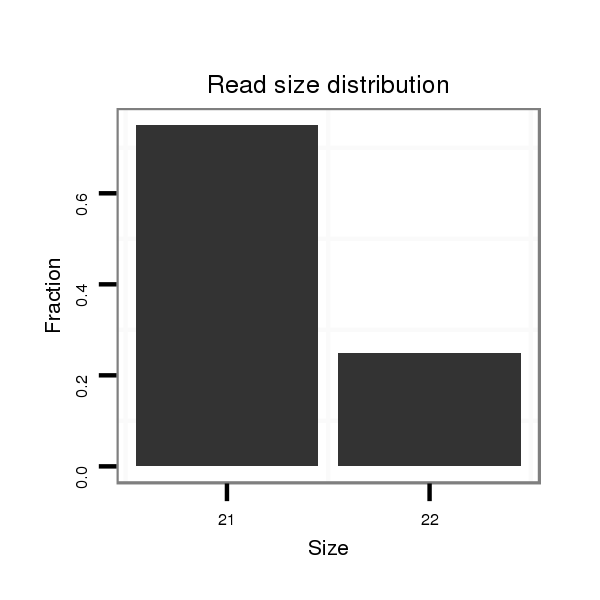
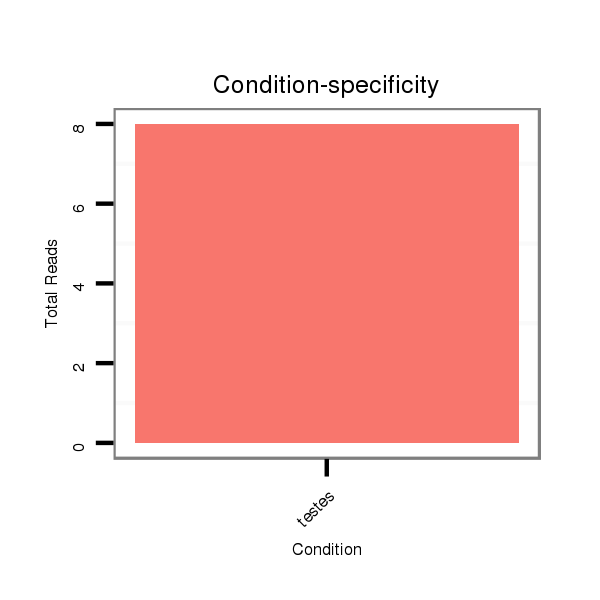
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR066810 testes |
SRR1274764 testes |
SRR1274763 ovary |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................ACCCTCATCCTGGTGTCCGGC................................................................................................. | 21 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 |
| ............................................................ACCCTCATCCTGGTGTCCAGC................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................TGTGTTTCTCTCCATGAAATAAGTCCCTC............... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................CCACTGTACAAACTTGCTGCCCACCTT..................................................................... | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................TAAGAGCTCAGAGTTCACCCTCATCC............................................................................................................ | 26 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ............................................................ACCCTCATCCTGGTGTCCGGCATT.............................................................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................CTGTGTTTCTCTCCATGAAATAAGTCCC................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................ACCCTCATCCTGGTGTCCGGCA................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CTGCCCACCTGGGTGAGAAGGA.......................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................TGCAAGTAAGAGCTCAGAGTTCACCCTCATC............................................................................................................. | 31 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................AAGTAAGAGCTCAGAGTTC...................................................................................................................... | 19 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................GTAGCACACACCCAGGCG................................................................................................................................................ | 18 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GTAAACCGGTGGGATCCATCGTGTGTGGGTCCGCTAGAACGTTCATTCTCGAGTCTCAAGTGGGAGTAGGACCACAGGCCGTGGTGACATGTTTGAACGACGGGTGGACCCACTCTTCCTGAGACACAAGGGAGACACAAAGAGAGGTACTTTATTCAGGGAGTACAACATTCTGAGA
*******************************************..((((.((((((((...(((((((.((((...((((.((..((....)).)).)))))))).)))))))..)))))))).)))).....********************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR066809 ovary |
|---|---|---|---|---|---|---|---|
| ......................................................CTCAAGTGGGAGTAGGACCA........................................................................................................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 |
| ............................................................................................................................CACAAGGGAGACACGAAGAGAGGTA............................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................................AGTCTCAAGTGGGAGTAGGAC.......................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| ................................................................................................................................TGGGAGACACAAAGAGAGGTA............................. | 21 | 1 | 7 | 0.29 | 2 | 2 | 0 |
Generated: 09/01/2015 at 11:34 AM