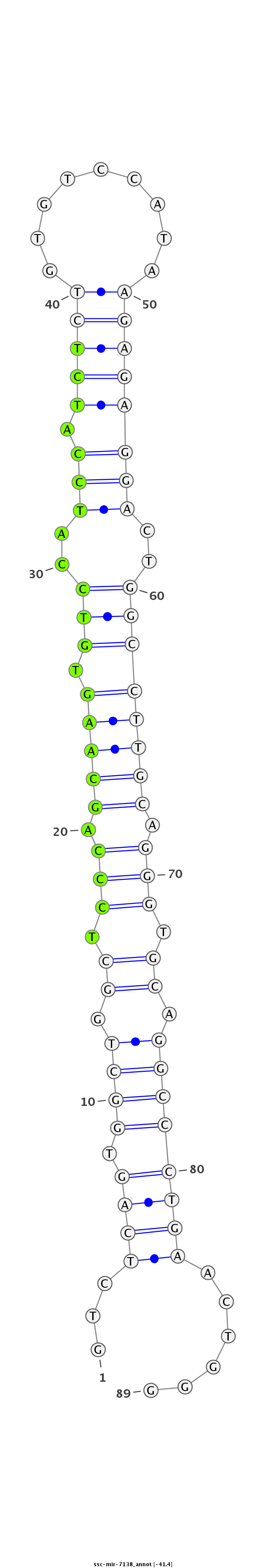


ID:ssc-mir-7138 |
Coordinate:chr2:82423677-82423735 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -41.1 | -41.0 |
 |
 |
|
GAGGAGGTGGCAGCGGGGCAGGACCCAACCATTGAGTCTCAGTGGCTGGCTCCCAGCAAGTGTCCATCCATCTCTGTGTCCATAAGAGAGGACTGGCCTTGCAGGGTGCAGGCCCTGAACTGGGCTGCAGAGCTGGCGGTGAGCCCCTTGGCTGTGTGT
***********************************...((((.((((.((.(((.(((((.(((..(((.(((((.........))))))))..)))))))).))).)).))))))))......*********************************** |
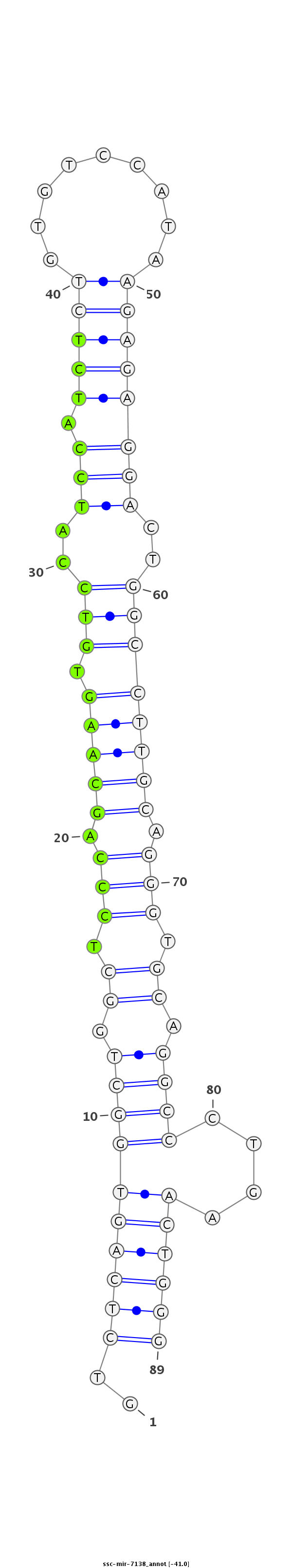
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR066809 ovary |
SRR1274763 ovary |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................TCCCAGCAAGTGTCCATCCATCT...................................................................................... | 23 | 0 | 1 | 17.00 | 17 | 14 | 2 | 1 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCCATC....................................................................................... | 22 | 0 | 1 | 5.00 | 5 | 3 | 2 | 0 | 0 |
| .......................................................................................GAGGACTGGCCTTGCAGGGTGT.................................................. | 22 | 1 | 1 | 4.00 | 4 | 3 | 0 | 1 | 0 |
| .......................................................................................GAGGACTGGCCTTGCAGGGTGTA................................................. | 23 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCCATCTC..................................................................................... | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ......................................................................................AGAGGACTGGCCTTGCAGGGTGTT................................................. | 24 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................AGAGGACTGGCCTTGCAGGGTGT.................................................. | 23 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 |
| .....................................................................................................................AACTGGGCTGCAGAGCTGGCGGTG.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................................AGAGGACTGGCCTTGCAGGGTGA.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCCATCA...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................GAGGACTGGCCTTGCAGGGTGCAA................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................GAGGACTGGCCTTGCAGGGTGA.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCCATCTCT.................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................GAGGACTGGCCTTGCAGGGTGCA................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................AGAGGACTGGCCTTGCAGGGTGTTT................................................ | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TCCCAGTAAGTGTCCATCCATCT...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCT.......................................................................................... | 19 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATCCAT........................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TCCCAGCAAGTGTCCATTCATCTTA.................................................................................... | 25 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 |
|
CTCCTCCACCGTCGCCCCGTCCTGGGTTGGTAACTCAGAGTCACCGACCGAGGGTCGTTCACAGGTAGGTAGAGACACAGGTATTCTCTCCTGACCGGAACGTCCCACGTCCGGGACTTGACCCGACGTCTCGACCGCCACTCGGGGAACCGACACACA
***********************************...((((.((((.((.(((.(((((.(((..(((.(((((.........))))))))..)))))))).))).)).))))))))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:32 AM