

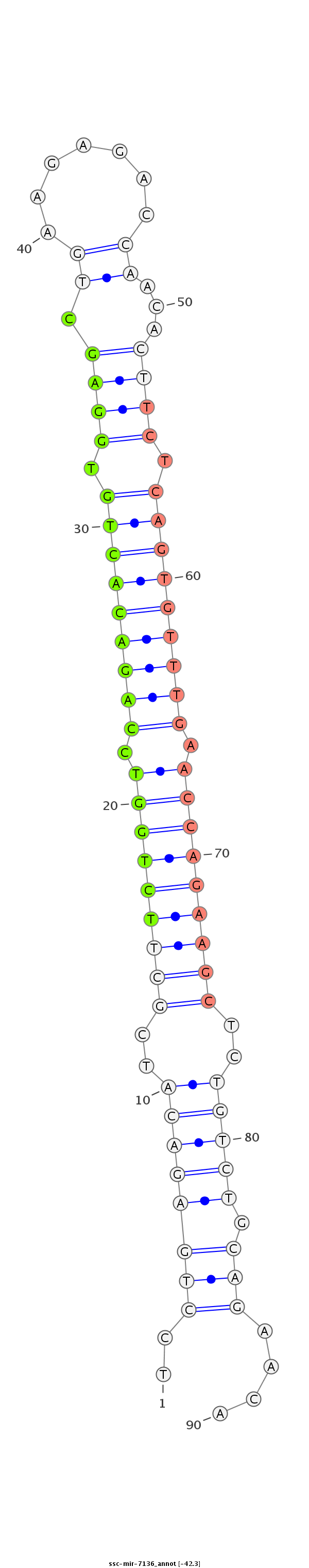
ID:ssc-mir-7136 |
Coordinate:chr7:8701439-8701498 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
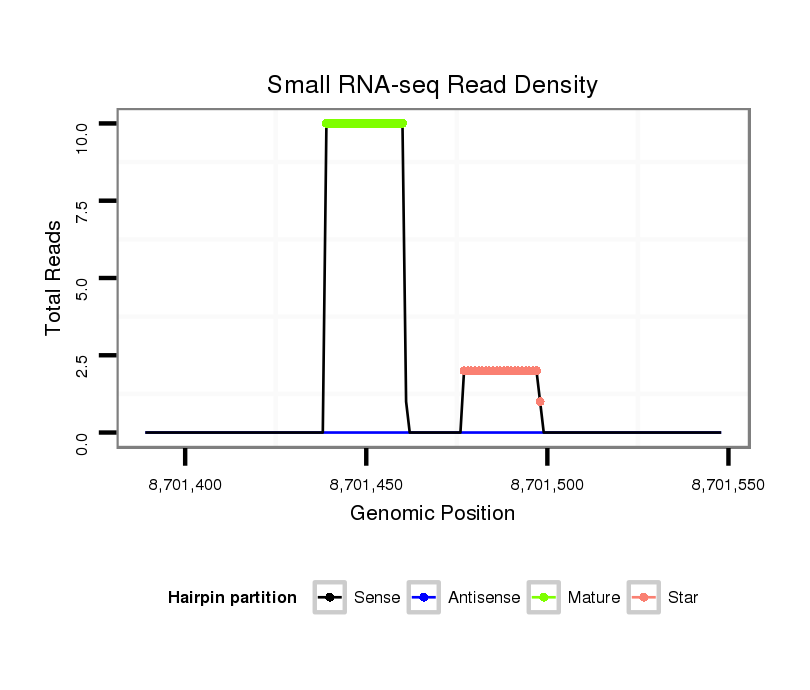
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -42.3 | -41.9 |
 |
 |
| mature | star |
|
TGTCTGTCTCCTTGGCCTCTCCAGACCATGCTGAGTCCTGAGACATCGCTTCTGGTCCAGACACTGTGGAGCTGAAGAGACCAACACTTCTCAGTGTTTGAACCAGAAGCTCTGTCTGCAGAACAATCAACAAAGCCGAGGAGCCCACATGCAACAGGAT
***********************************..((((((((..(((((((((.(((((((((.((((.((..........)))))).))))))))).)))))))))..))))).)))....*********************************** |
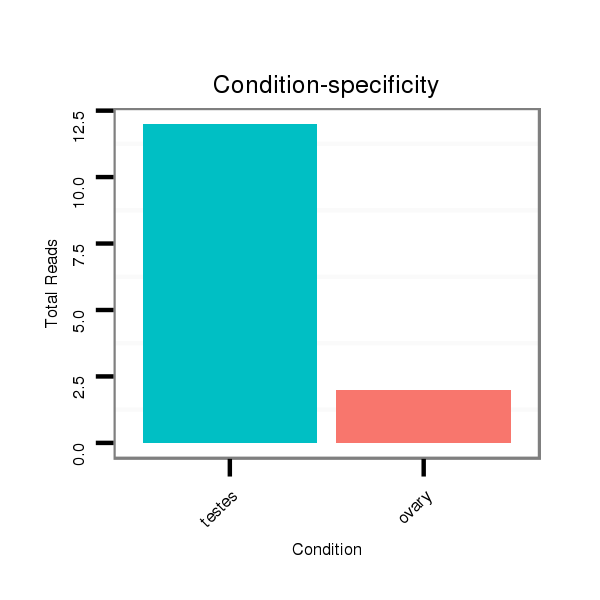
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066810 testes |
SRR139075 testes |
SRR1274764 testes |
SRR066809 ovary |
SRR139076 testes |
SRR1274763 ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TCTGGTCCAGACACTGTGGAGC........................................................................................ | 22 | 0 | 2 | 9.00 | 18 | 4 | 3 | 7 | 2 | 1 | 1 |
| ..................................................TCTGGTCCAGACACTGTGGAGCT....................................................................................... | 23 | 0 | 2 | 1.50 | 3 | 0 | 2 | 1 | 0 | 0 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAGT.................................................. | 22 | 1 | 2 | 1.50 | 3 | 0 | 1 | 0 | 0 | 2 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAGC.................................................. | 22 | 0 | 2 | 1.50 | 3 | 2 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAG................................................... | 21 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAGA.................................................. | 22 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TCTGGTCCAGACACTGTGGAGA........................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCTGGTCCAGACACTGCGGAGA........................................................................................ | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TTCTCAGTGTTTGAACCAGAAGA.................................................. | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCTGGTCCAGACATTGTGGAGC........................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCTGGTCCAGACACTGTGGAGCTG...................................................................................... | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TTCTCAGTGTTTGAACCAGAAGTA................................................. | 24 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TCTGGTCCAGACACTGTGGAGT........................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................CTTCTGGTCCAGACACTGTG............................................................................................ | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TTCTCAGTGTTTGAACCAGA..................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCAGTGTTTGAACCAGAAGCTC................................................ | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAT................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCTCAGTGTTTGAACCAGAAGTA................................................. | 23 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACAGACAGAGGAACCGGAGAGGTCTGGTACGACTCAGGACTCTGTAGCGAAGACCAGGTCTGTGACACCTCGACTTCTCTGGTTGTGAAGAGTCACAAACTTGGTCTTCGAGACAGACGTCTTGTTAGTTGTTTCGGCTCCTCGGGTGTACGTTGTCCTA
***********************************..((((((((..(((((((((.(((((((((.((((.((..........)))))).))))))))).)))))))))..))))).)))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR139075 testes |
|---|---|---|---|---|---|---|---|
| ..............................................GCGAAGACCAGGTCTGTGACAC............................................................................................ | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ......................................TCTCTGTAGCGAAGACCAGGTCT................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 1 |
| .............................................AGCGAAGACCAGGTCTGTGAC.............................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ...............................................CGAAGACCAGGTCTGTGACACC........................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 |
Generated: 09/01/2015 at 11:41 AM