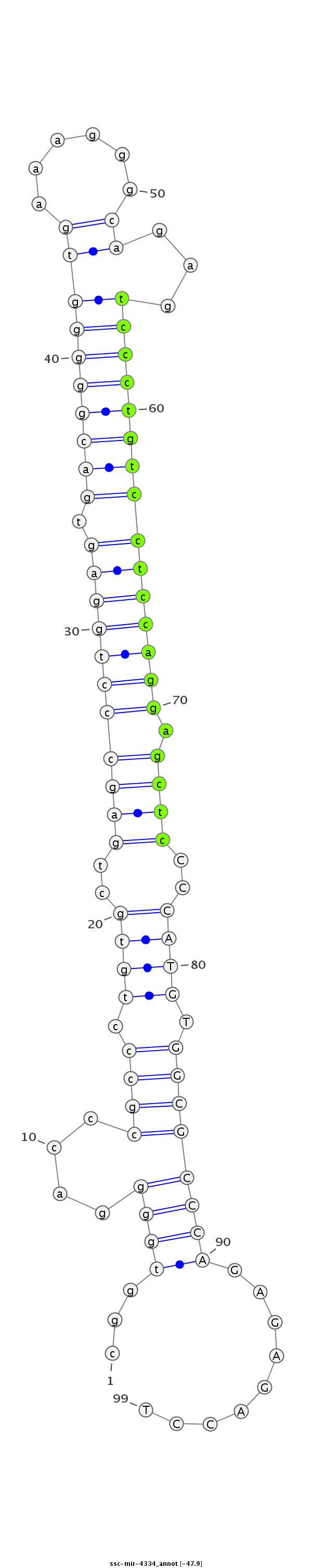
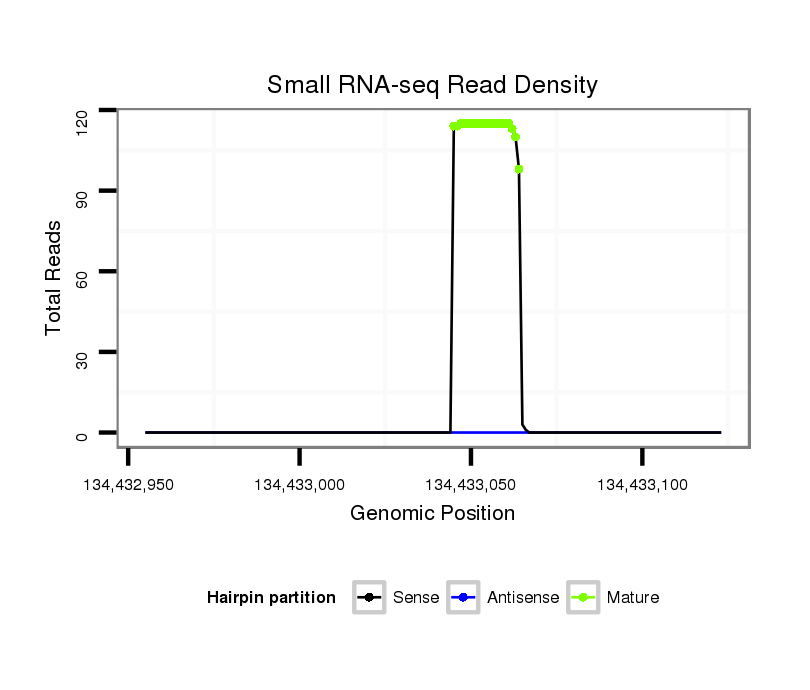
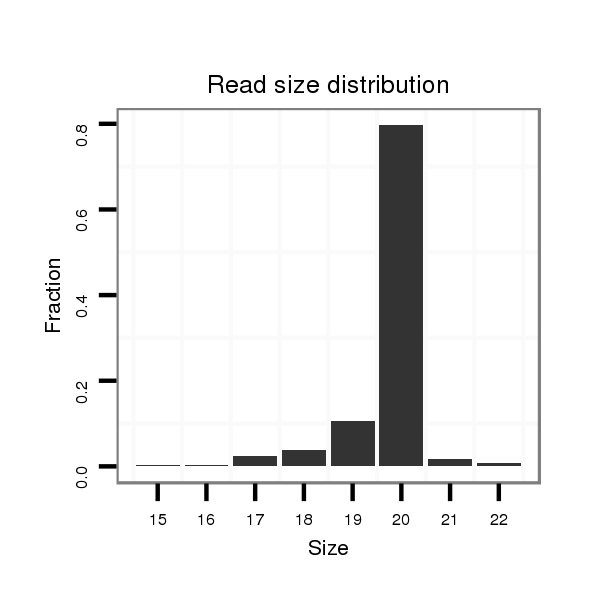
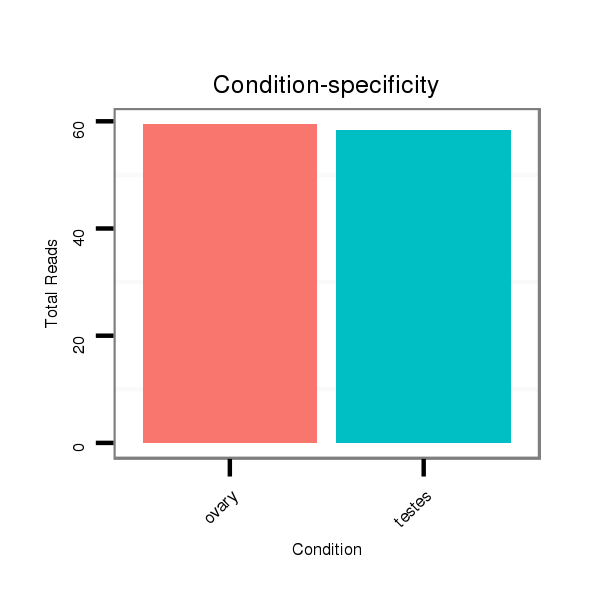
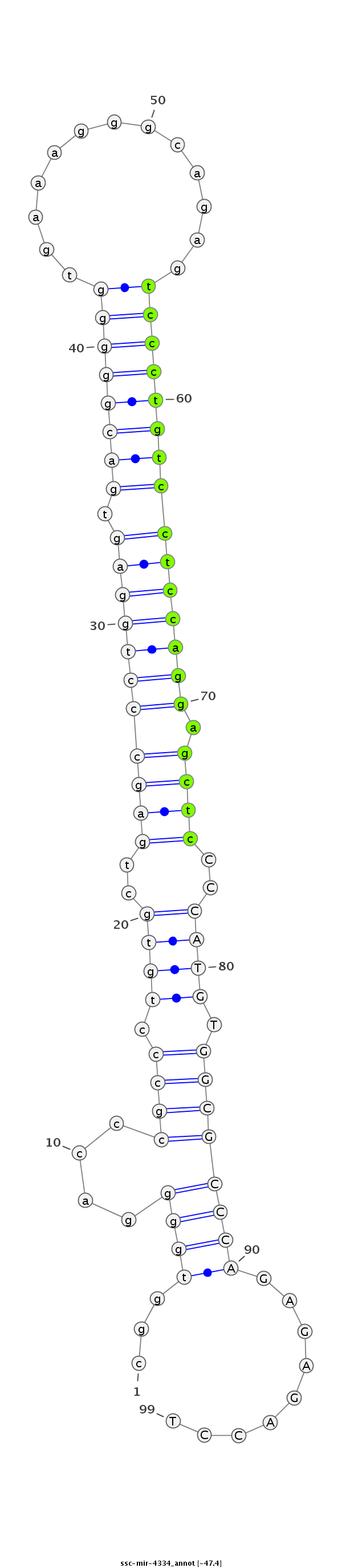
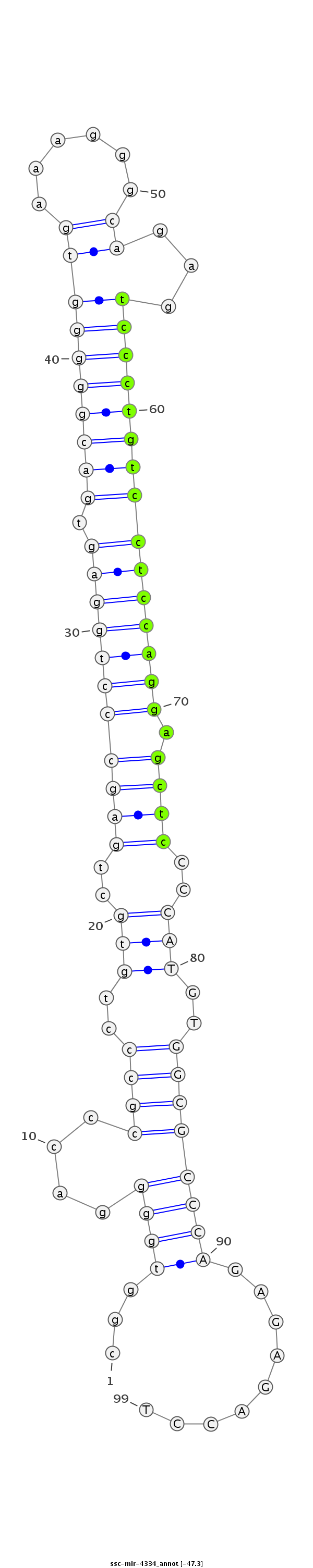
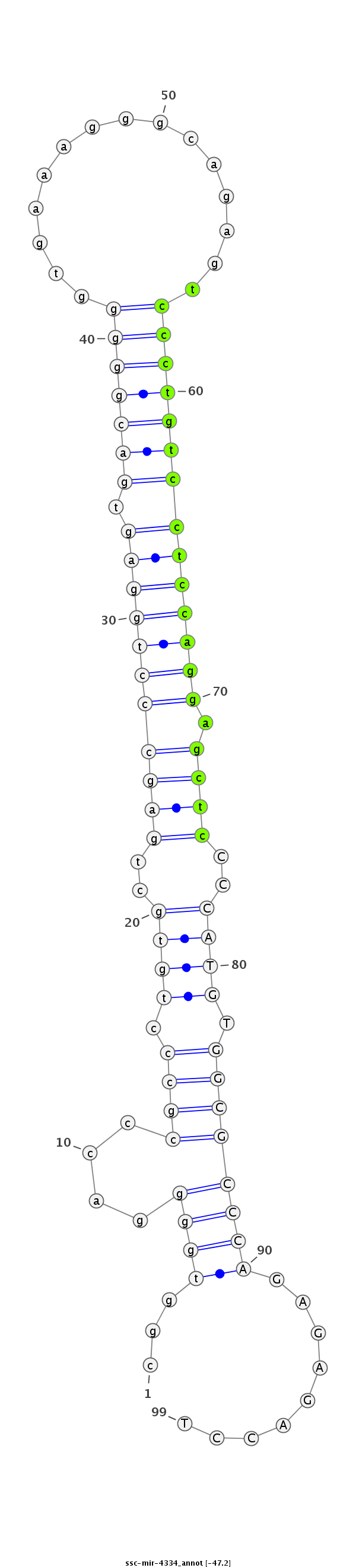
ID:ssc-mir-4334 |
Coordinate:chr15:134433005-134433073 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -47.4 | -47.3 | -47.2 |
 |
 |
 |
| mature | star |
|
CCGAGTCTAAGCTGGCACCCTCACTtaagcacgtgcggtggggacccgccctgtgctgagccctggagtgacgggggtgaaagggcagagtccctgtcctccaggagctcCCCATGTGGCGCCCAGAGAGACCTGGGAGGCGTGGTCATGCTAGGAGCAGACACACCGA
***********************************...((((....((((.((((..(((((((((((.((((((((((......))...))))))))))))))).))))..)))).)))))))).........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1274763 ovary |
SRR139075 testes |
SRR066809 ovary |
SRR1274764 testes |
SRR066810 testes |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................TCCCTGTCCTCCAGGAGCTC........................................................... | 20 | 0 | 6 | 94.00 | 564 | 260 | 134 | 74 | 65 | 30 | 1 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCT............................................................ | 19 | 0 | 11 | 12.64 | 139 | 10 | 97 | 21 | 1 | 5 | 5 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCACC........................................................ | 23 | 1 | 4 | 4.75 | 19 | 4 | 7 | 1 | 0 | 7 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGC............................................................. | 18 | 0 | 14 | 3.36 | 47 | 1 | 36 | 6 | 0 | 3 | 1 |
| ..........................................................................................TCCCTGTCCTCCAGGAG.............................................................. | 17 | 0 | 17 | 2.76 | 47 | 0 | 46 | 0 | 0 | 0 | 1 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCACCA....................................................... | 24 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCC.......................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................CCTGTCCTCCAGGAGCTC........................................................... | 18 | 0 | 7 | 1.29 | 9 | 2 | 6 | 1 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCCC......................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTA........................................................... | 20 | 1 | 16 | 0.69 | 11 | 1 | 6 | 2 | 0 | 2 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCG.......................................................... | 21 | 1 | 6 | 0.67 | 4 | 1 | 1 | 1 | 0 | 1 | 0 |
| ..........................................................................................TCTCTGTCCTCCAGGAGCTC........................................................... | 20 | 1 | 19 | 0.47 | 9 | 0 | 9 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGA............................................................. | 18 | 1 | 20 | 0.30 | 6 | 0 | 0 | 1 | 0 | 5 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGA............................................................... | 16 | 0 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGG................................................................ | 15 | 0 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 |
| ............................................................................................CCTGTCCTCCAGGAGCTCACTAT...................................................... | 23 | 2 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCACCAGT..................................................... | 26 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCACCAGGAGCTC........................................................... | 20 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCTCGA......................................................... | 22 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTACTCCAGGAGCTC........................................................... | 20 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTCTCCTCCAGGAGCTC........................................................... | 20 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTCCCTGTCCTCCAGGAGCTC........................................................... | 21 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................CCTGTCCTCCAGGAGCT............................................................ | 17 | 0 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCAGTCCTCCAGGAGCTC........................................................... | 20 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TTCCTGTCCTCCAGGAGCTC........................................................... | 20 | 1 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................CCTGTCCTCCAGGAGCTCATCA....................................................... | 22 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ..........................................................................................TCTCTGTCCTCCAGGAGCT............................................................ | 19 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................CTGTCCTCCAGGAGCTC........................................................... | 17 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGTTC........................................................... | 20 | 1 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCGGGAGCTC........................................................... | 20 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCACGAGCT............................................................ | 19 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................CTGTCCTCCAGGAGCT............................................................ | 16 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCCGTCCTCCAGGAG.............................................................. | 17 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCTGTCCTCCAGGAGCA............................................................ | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................CCTGTCCTCCAGGAGC............................................................. | 16 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GGCTCAGATTCGACCGTGGGAGTGAtTTgGTGgtgGggtggggTGGGgGGGtgtgGtgTgGGGtggTgtgTGgggggtgTTTgggGTgTgtGGGtgtGGtGGTggTgGtGGGGTACACCGCGGGTCTCTCTGGACCCTCCGCACCAGTACGATCCTCGTCTGTGTGGCT
***********************************...((((....((((.((((..(((((((((((.((((((((((......))...))))))))))))))).))))..)))).)))))))).........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:28 AM