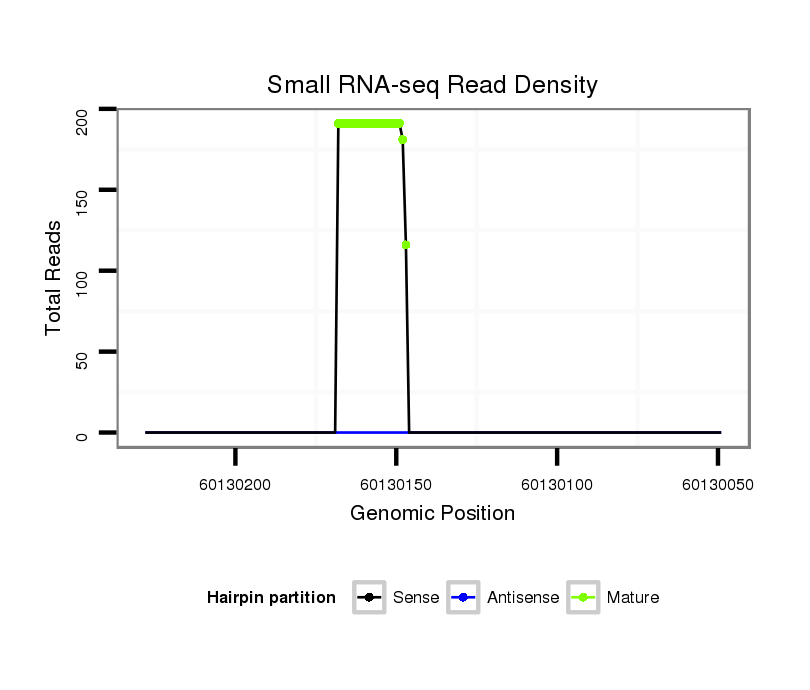

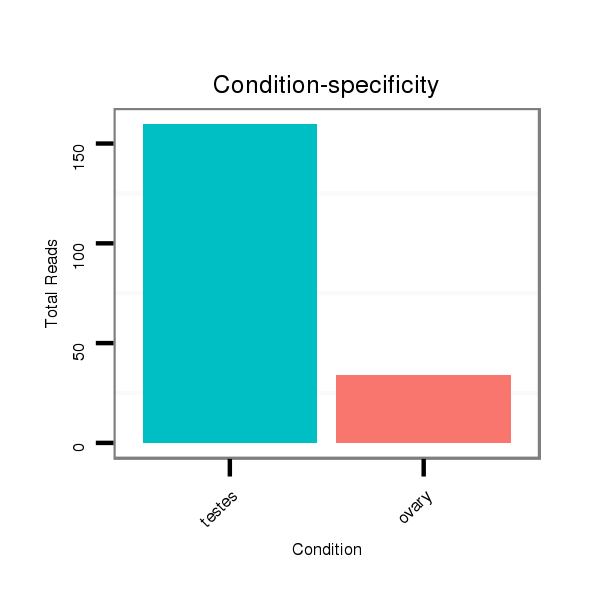
ID:ssc-mir-4331 |
Coordinate:chr6:60130099-60130178 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| mature | star |
|
tgGgtGGTTggtTgggTGGggTGGgTTtGTGGGTgtGGGtTggtGgtTTGggtTGtGgTtTGGgGTtGtTgtgtGtgtgtGgTgtGtTgTGGgtTTGgTGTGGgTGTGGTGTtGGggtGgtGgTTTGgTggttTTgttggTgTGGggTGGGTGtGGgggTtttttgtttttttGttGCCC
***********************************..............................................................................................................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR1274763 ovary |
SRR1274764 testes |
SRR139076 testes |
SRR066809 ovary |
SRR066810 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TGGCGTAGATCACAGACACAGC.................................................................................................. | 22 | 0 | 2 | 116.50 | 233 | 145 | 29 | 26 | 12 | 16 | 5 |
| ............................................................TGGCGTAGATCACAGACACAG................................................................................................... | 21 | 0 | 2 | 65.00 | 130 | 83 | 15 | 20 | 4 | 5 | 3 |
| ............................................................TGGCGTAGATCACAGACACAGCA................................................................................................. | 23 | 1 | 5 | 11.00 | 55 | 51 | 0 | 0 | 3 | 1 | 0 |
| ............................................................TGGCGTAGATCACAGACACA.................................................................................................... | 20 | 0 | 2 | 10.00 | 20 | 12 | 3 | 0 | 3 | 0 | 2 |
| ............................................................TGGCGTAGATCACAGACACAGA.................................................................................................. | 22 | 1 | 14 | 1.57 | 22 | 18 | 0 | 1 | 3 | 0 | 0 |
| ............................................................TGGCGTAGATCACCGACACAGC.................................................................................................. | 22 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................GGCGTAGATCACAGACACAGC.................................................................................................. | 21 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GGCGTAGATCACAGACACAG................................................................................................... | 20 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACCGACACAG................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACACACACAG................................................................................................... | 21 | 1 | 4 | 0.50 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGCT................................................................................................. | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACAC..................................................................................................... | 19 | 0 | 8 | 0.38 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGCG................................................................................................. | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGAA................................................................................................. | 23 | 2 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................GCGTAGATCACCGACACAGC.................................................................................................. | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GGCGTAGATCACGGACACAGC.................................................................................................. | 21 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAA................................................................................................... | 21 | 1 | 19 | 0.16 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GGCGTAGATCACAGACACAGCA................................................................................................. | 22 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................CGTAGATCACAGACACAG................................................................................................... | 18 | 0 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGCC................................................................................................. | 23 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAT................................................................................................... | 21 | 1 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGGCACAG................................................................................................... | 21 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGCAG................................................................................................ | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAAACACAGC.................................................................................................. | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACAGT.................................................................................................. | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TGGCGTAGATCACAGACACCGCA................................................................................................. | 23 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GGCGTAGATCACAGACACAGT.................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
tgCgtCCAAggtAgggACCggACCgAAtCACCCAgtCCCtAggtCgtAACggtACtCgAtACCgCAtCtAgtgtCtgtgtCgAgtCtAgACCgtAACgACACCgACACCACAtCCggtCgtCgAAACgAggttAAgttggAgACCggACCCACtCCgggAtttttgtttttttCttCGGG
***********************************..............................................................................................................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:39 AM