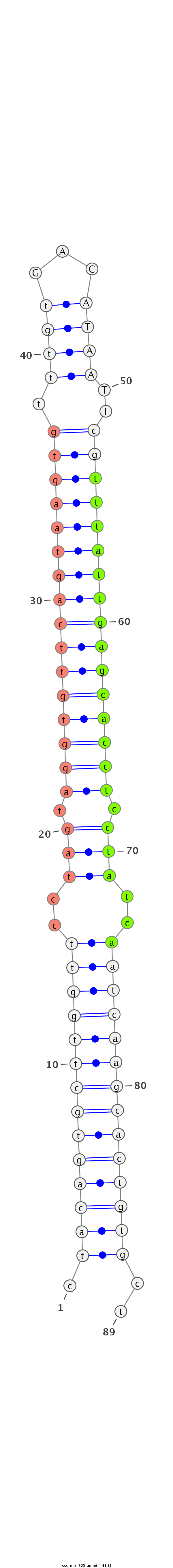
ID:ssc-mir-325 |
Coordinate:chrX:69436727-69436820 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.7 | -40.6 | -40.4 |
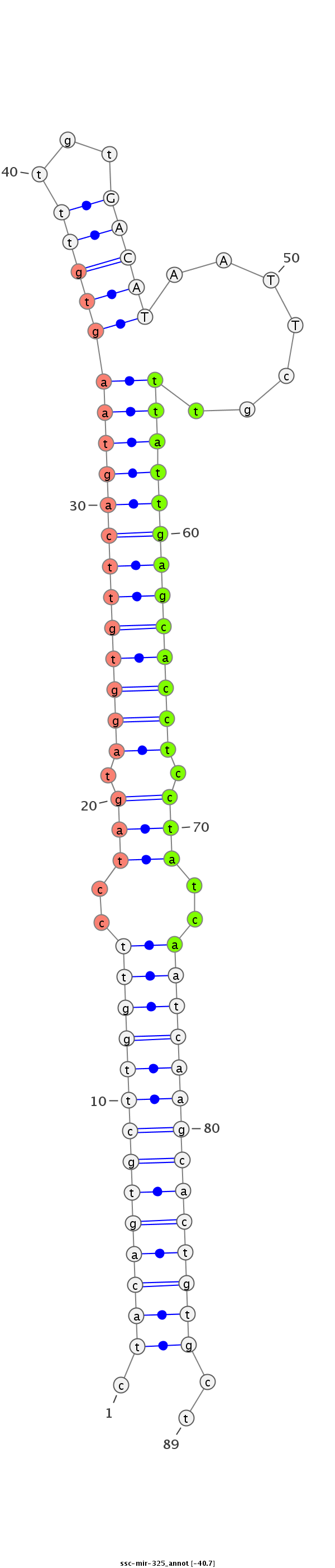 |
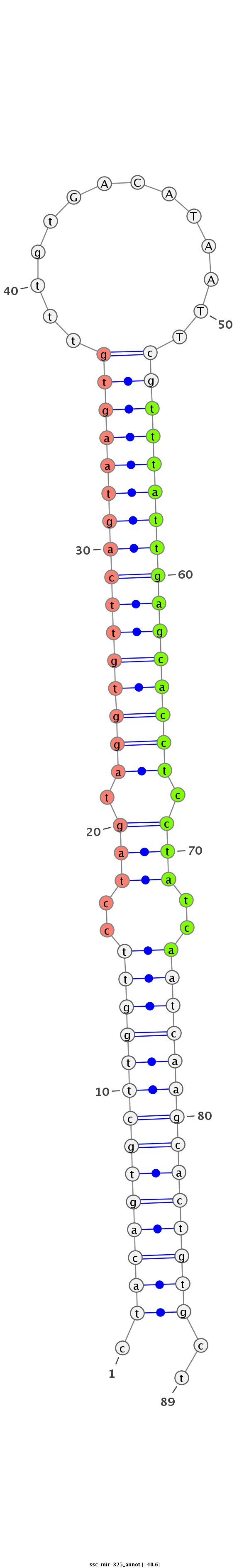 |
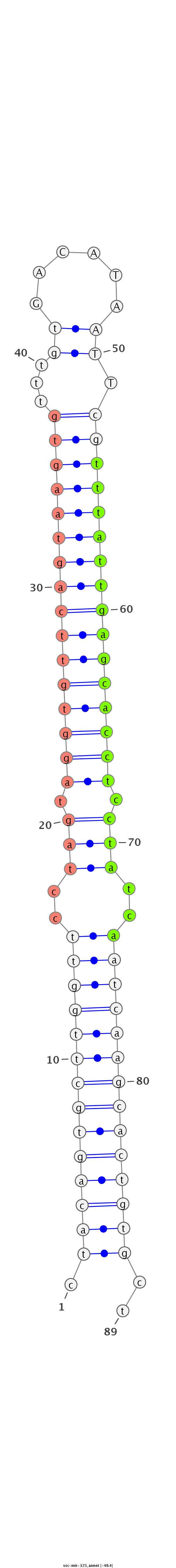 |
| mature | star |
|
GTGATAGATTACAGGGTTCCAAGTGTTGTGGATTCTAGcagagccgctacagtgcttggttcctagtaggtgttcagtaagtgtttgtGACATAATTcgtttattgagcacctcctatcaatcaagcactgtgctaggttctgggaccacacagataagatagacaaggtcctacttttcgaggtgctcatgtt
**********************************************.((((((((((((((..(((.((((((((((((((((.((((...))))..)))))))))))))))).)))..))))))))))))))..*********************************************************** |
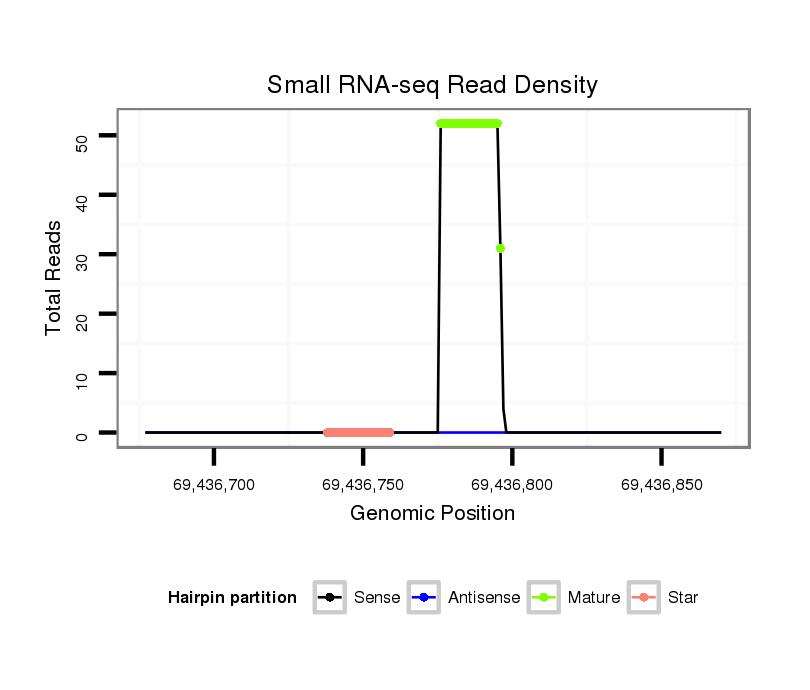
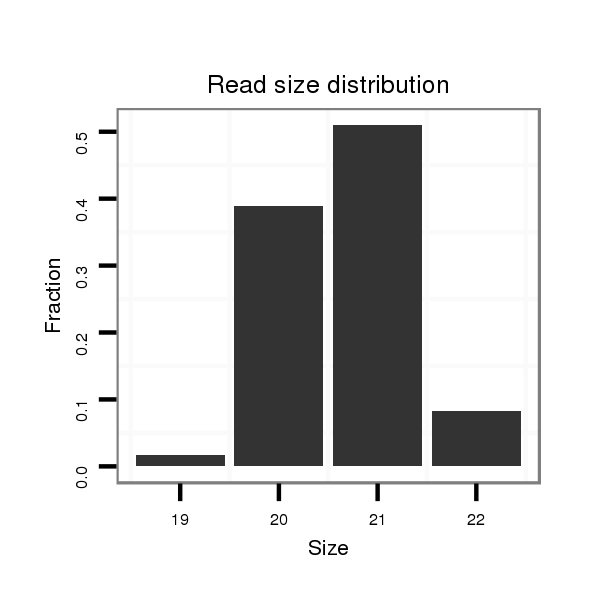
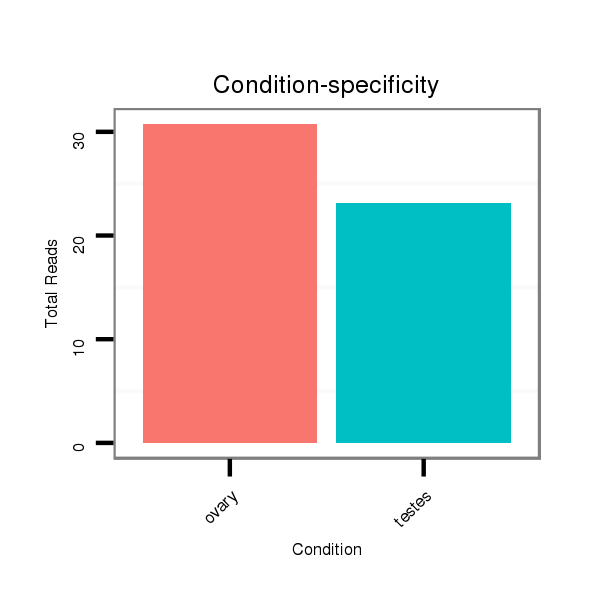
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR066810 testes |
SRR139075 testes |
|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TTTATTGAGCACCTCCTATCA.......................................................................... | 21 | 0 | 2 | 27.50 | 55 | 31 | 24 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTATC........................................................................... | 20 | 0 | 2 | 21.00 | 42 | 26 | 16 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTATCAA......................................................................... | 22 | 0 | 2 | 4.00 | 8 | 4 | 4 | 0 |
| .............................................................CCTAGTAGGTGCTCAGTAAGTGT.............................................................................................................. | 23 | 1 | 8 | 0.88 | 7 | 6 | 1 | 0 |
| .....................................................................................................TATTGAGCACCTCCTATCA.......................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .........................................................................................................................TCAAGCACTGTGCTAGGCTCTGGGGCCACATA......................................... | 32 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .............................................................CCTAGTAGGTGTTCAGTAAGTG............................................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| ...................................................................................................TTTATTGAGCACCTCCTATCAAG........................................................................ | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTATCAAGTAG..................................................................... | 26 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTAT............................................................................ | 19 | 0 | 19 | 0.42 | 8 | 5 | 3 | 0 |
| .............................................................CCTAGTAGGTGCTCAGTAAGTG............................................................................................................... | 22 | 1 | 17 | 0.41 | 7 | 6 | 1 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTATCAT......................................................................... | 22 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...................................................................................................TTTATTGAGCACCTCCTATCG.......................................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ..................................................................................................GTTTATTGAGCACCTCCTATCT.......................................................................... | 22 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| .............................................................CCTAGTAGGTGCTCAGTAAGT................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ........................................................................................................TGAGCACCGCCTATCGA......................................................................... | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
CACTATCTAATGTCCCAAGGTTCACAACACCTAAGATCGTgTgGGgGtTGTgtgGttggttGGtTgtTggtgttGTgtTTgtgtttgtCTGTATTAAGgtttTttgTgGTGGtGGtTtGTTtGTTgGTGtgtgGtTggttGtgggTGGTGTGTgTtTTgTtTgTGTTggtGGtTGttttGgTggtgGtGTtgtt
***********************************************************.((((((((((((((..(((.((((((((((((((((.((((...))))..)))))))))))))))).)))..))))))))))))))..********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:45 AM