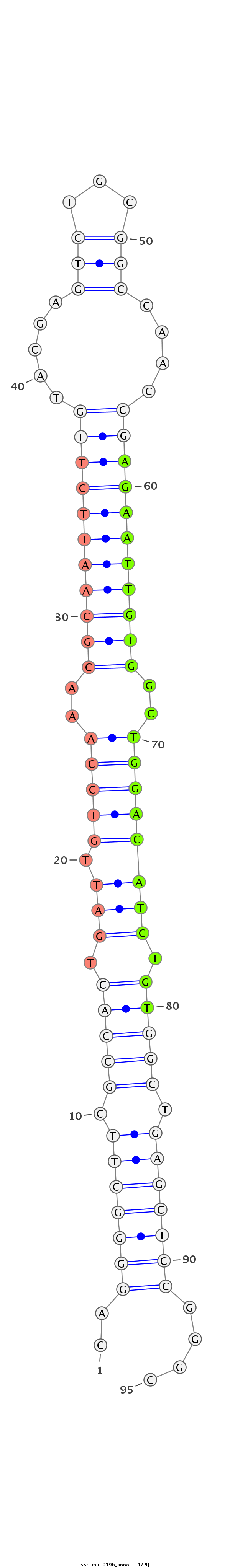
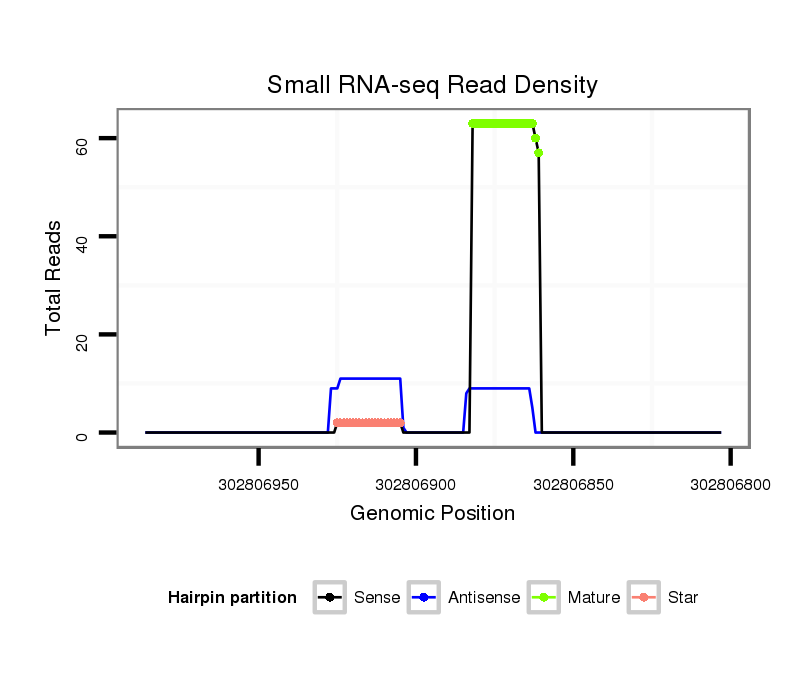
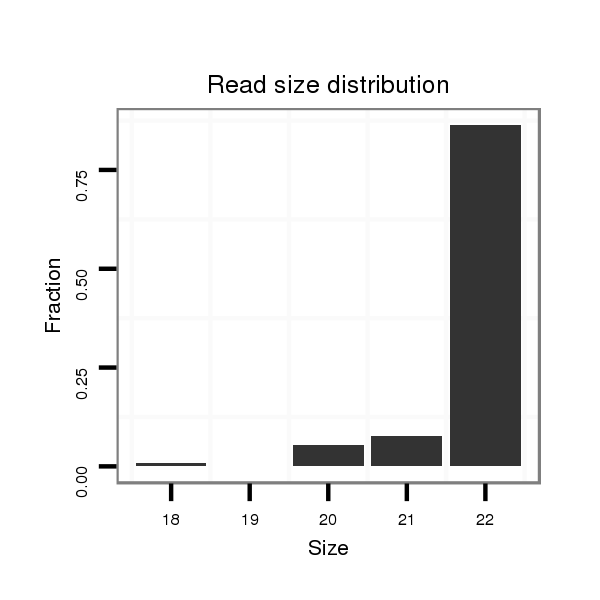
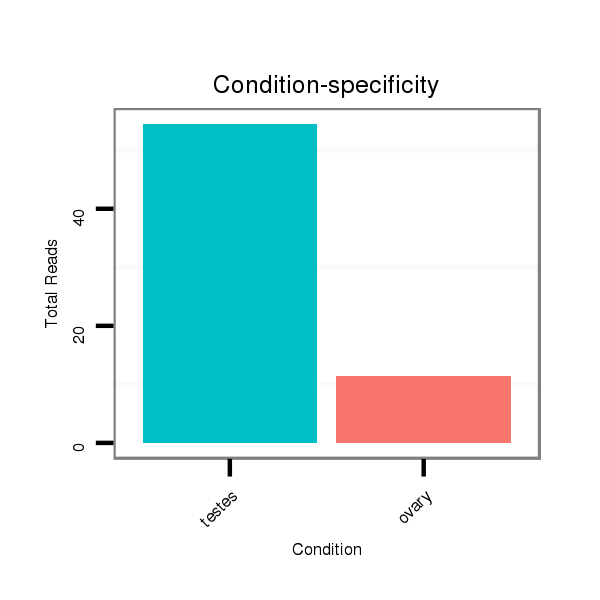
ID:ssc-mir-219b |
Coordinate:chr1:302806853-302806936 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
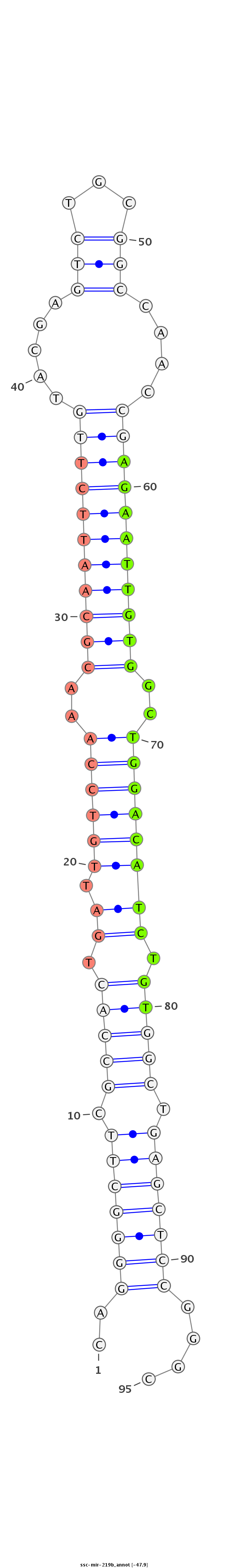
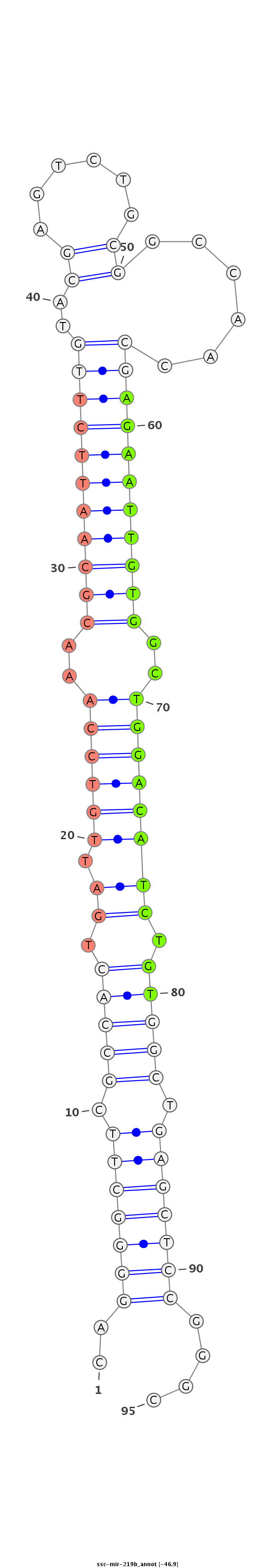
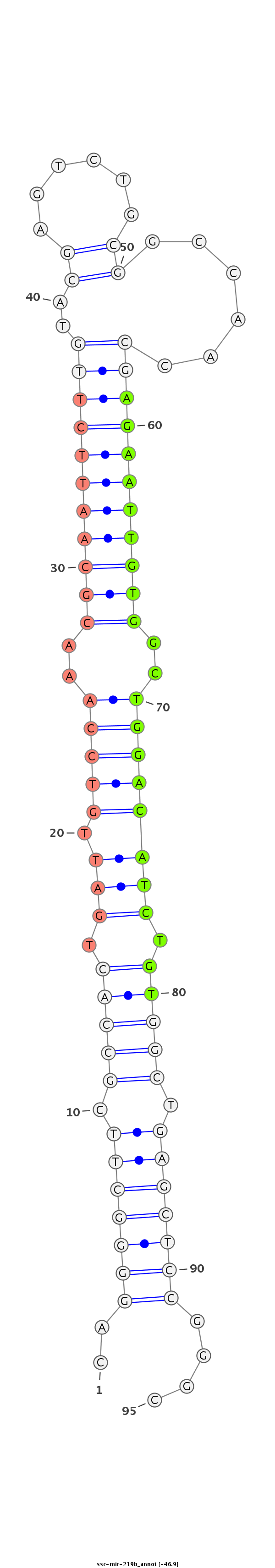
| -47.9 | -46.9 | -46.9 |
 |
 |
 |
| mature | star |
|
TTTAAGGAGCGAAGCGGAACCCCGCAGGAGACTGGGGCCCTTAACTCAGGGGCTTCGCCACTGATTGTCCAAACGCAATTCTTGTACGAGTCTGCGGCCAACCGAGAATTGTGGCTGGACATCTGTGGCTGAGCTCCGGGCGCAACGGGGGCGGGAGTCGCAGGGACCGAGCTCAGCGCGGGAG
**********************************************..(((((((.(((((.(((.(((((..(((((((((((.....(((...)))....)))))))))))..)))))))).))))).)))))))....******************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139076 testes |
SRR066809 ovary |
SRR066810 testes |
SRR139075 testes |
SRR1274763 ovary |
SRR1274764 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................AGAATTGTGGCTGGACATCTGT.......................................................... | 22 | 0 | 1 | 57.00 | 57 | 37 | 10 | 9 | 1 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCTGA.......................................................... | 22 | 1 | 1 | 6.00 | 6 | 5 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCT............................................................ | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCTG........................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCTGTAA........................................................ | 24 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCTGTAT........................................................ | 24 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................TGATTGTCCAAACGCAATTCT...................................................................................................... | 21 | 0 | 2 | 2.00 | 4 | 1 | 1 | 0 | 2 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACATCTGTT......................................................... | 23 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| .............................................................TGATTGTCCAAACGCAATTCTCGT................................................................................................... | 24 | 1 | 2 | 2.00 | 4 | 0 | 1 | 1 | 0 | 1 | 1 |
| ........................................................................................................AGAATTGTGGCTGGACATCTGG.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTCGACCTCTGT.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTCGACATCT............................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGACCTCTGT.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGTGGCTGGGCATCTGT.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTGGGGCTGGACATCTGT.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGACTTGTGGCTGGACATCTGT.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ATTGCGGCTGGACATCTGAG......................................................... | 20 | 2 | 4 | 0.75 | 3 | 0 | 2 | 1 | 0 | 0 | 0 |
| .............................................................TGATTGTCAAAACGCAATTCT...................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................TGATTGTCCAAACGTAATT........................................................................................................ | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................TGATTGTCCAAACGCAATTC....................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................TGATTGTCCAAACGTAATTC....................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................TGATTGTCCAAACGCAAT......................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................ATTGCGGCTGGACATCTGA.......................................................... | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................GGGCGCAACGGGTGCCGG............................. | 18 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
AAATTCCTCGCTTCGCCTTGGGGCGTCCTCTGACCCCGGGAATTGAGTCCCCGAAGCGGTGACTAACAGGTTTGCGTTAAGAACATGCTCAGACGCCGGTTGGCTCTTAACACCGACCTGTAGACACCGACTCGAGGCCCGCGTTGCCCCCGCCCTCAGCGTCCCTGGCTCGAGTCGCGCCCTC
*******************************************..(((((((.(((((.(((.(((((..(((((((((((.....(((...)))....)))))))))))..)))))))).))))).)))))))....********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR066809 ovary |
SRR066810 testes |
|---|---|---|---|---|---|---|---|---|
| ...........................................................TGACTAACAGGTTTGCGTTAAGA...................................................................................................... | 23 | 0 | 1 | 9.00 | 9 | 2 | 4 | 3 |
| ......................................................................................................GCTCTTAACACCGACCTGTAG............................................................. | 21 | 0 | 1 | 4.00 | 4 | 0 | 3 | 1 |
| ......................................................................................................GCTCTTAACACCGACCTGTAGA............................................................ | 22 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 |
| .....................................................................................................AGCTCTTAACACCGACCTGTAGA............................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| .......................................................................................................CTCTTAACACCGACCTGTAGA............................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................CTAACAGGTTTGCGTTAAGA...................................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 0 | 1 | 1 |
| .......................................................................................................CTCTTAACACCGACCTGTCGA............................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................CTAACAGGTTTGCGTTAAGAA..................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ................................................................................................................................GACTCGATGCCCGCG......................................... | 15 | 1 | 20 | 0.10 | 2 | 0 | 0 | 2 |
| ...............................................................................................................................CGACTCGATGCCCGCG......................................... | 16 | 1 | 13 | 0.08 | 1 | 0 | 0 | 1 |
Generated: 09/01/2015 at 11:22 AM