
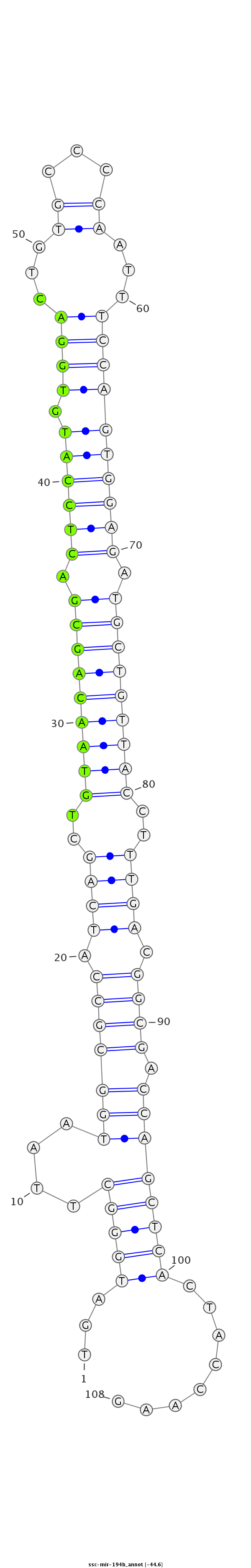
ID:ssc-mir-194b |
Coordinate:chr10:11962863-11962940 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
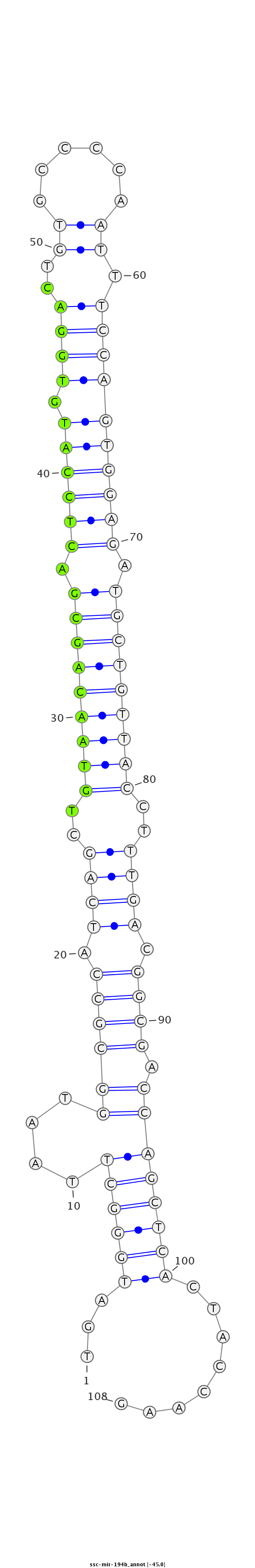
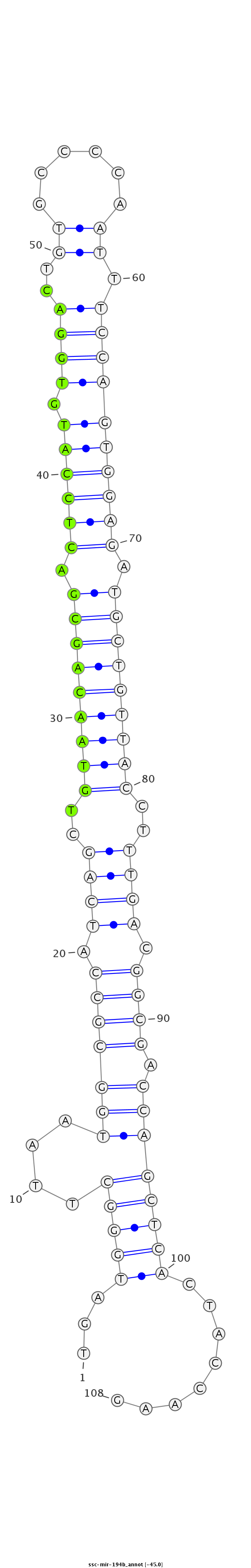
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -45.0 | -44.6 | -44.6 |
 |
 |
 |
| mature | star |
|
ATAGGCTTTCATCGAGACGAGCGGAAGGAGCCCTCTGATGGGCTTAATGGCGCCATCAGCTGTAACAGCGACTCCATGTGGACTGTGCCCCAATTTCCAGTGGAGATGCTGTTACCTTTGACGGCGACCAGCTCACTACCAAGATCCAAGAAGGACAATAAAAAAAATCATGGTCACT
***********************************...((((((....((((((.((((..(((((((((.((((((.((((..((......)).)))))))))).)))))))))..)))).)))).))))))))........*********************************** |
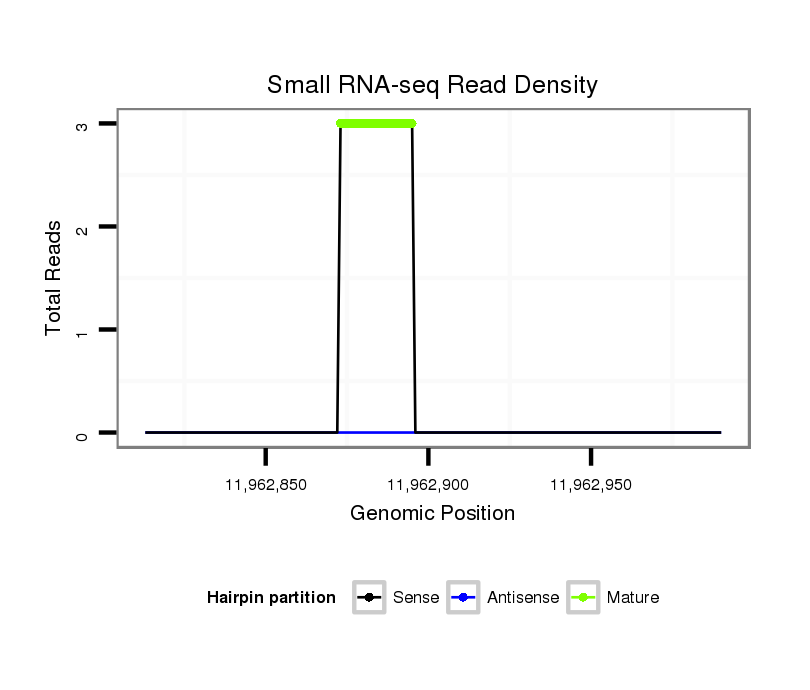
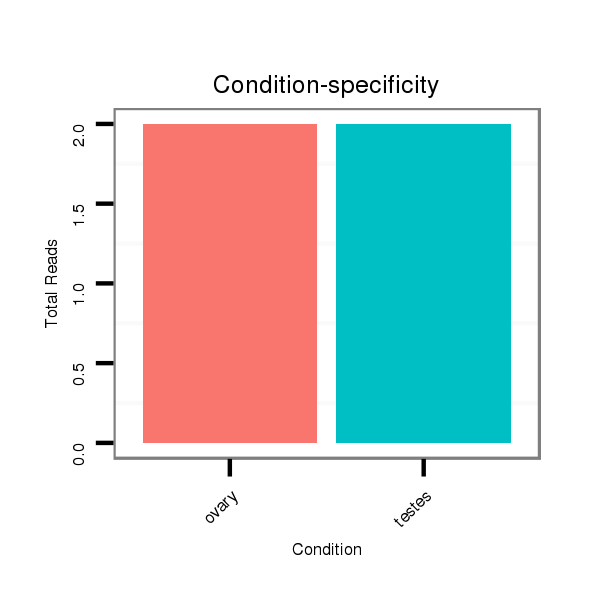
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR066810 testes |
SRR139075 testes |
SRR1274763 ovary |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................TGTAACAGCAACTCCATGTGGAC............................................................................................... | 23 | 1 | 3 | 29.67 | 89 | 48 | 41 | 0 | 0 |
| .........................................................ATATGTAACAGCAACTCCATGTGG................................................................................................. | 24 | 3 | 4 | 4.75 | 19 | 18 | 1 | 0 | 0 |
| ............................................................TGTAACAGCGACTCCATGTGGAC............................................................................................... | 23 | 0 | 2 | 3.50 | 7 | 3 | 4 | 0 | 0 |
| ............................................................TGTAACAGCGACTCCATGTGGACA.............................................................................................. | 24 | 1 | 2 | 2.00 | 4 | 0 | 0 | 3 | 1 |
| ............................................................TGTAACAGCAACTCCATGTGGACA.............................................................................................. | 24 | 2 | 4 | 1.75 | 7 | 4 | 3 | 0 | 0 |
| ............................................................TGTAACAGCAACTCCATGTGGACT.............................................................................................. | 24 | 1 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 |
| .........................................................ATATGTAACAGCAACTCCATGTGGA................................................................................................ | 25 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 |
| ............................................................TGTAACAGCAACTCCATGTGGATT.............................................................................................. | 24 | 2 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 |
| ............................................................TGTAACAGCGACTCCATGTGGA................................................................................................ | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................ATTTGTAACAGCAACTCCATGTGG................................................................................................. | 24 | 3 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 |
| ............................................................TGTAACAGCAACTCTATGTGGAC............................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ............................................................CGTAACAGCAACTCCATGTGGAC............................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................................................TGTAACAGCAACTCCCTGTGGAC............................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................................................TGTAACAGCAACTCCATGTGGAGT.............................................................................................. | 24 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................................................TGTAACATCAACTCCATGTGGAC............................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ............................................................TGTAACAGCAATTCCATGTGGAC............................................................................................... | 23 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .........................................................AAATGTAACAGCAACTCCATGTGGA................................................................................................ | 25 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ............................................................TGTAACAGCAACTCCATGTGGCC............................................................................................... | 23 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
|
TATCCGAAAGTAGCTCTGCTCGCCTTCCTCGGGAGACTACCCGAATTACCGCGGTAGTCGACATTGTCGCTGAGGTACACCTGACACGGGGTTAAAGGTCACCTCTACGACAATGGAAACTGCCGCTGGTCGAGTGATGGTTCTAGGTTCTTCCTGTTATTTTTTTTAGTACCAGTGA
***********************************...((((((....((((((.((((..(((((((((.((((((.((((..((......)).)))))))))).)))))))))..)))).)))).))))))))........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:23 AM