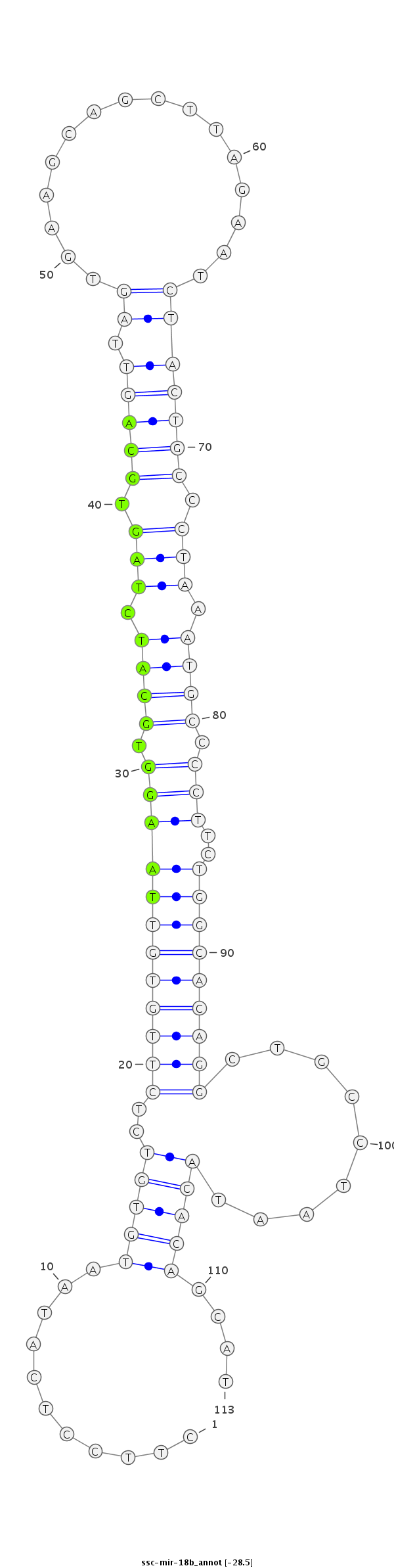


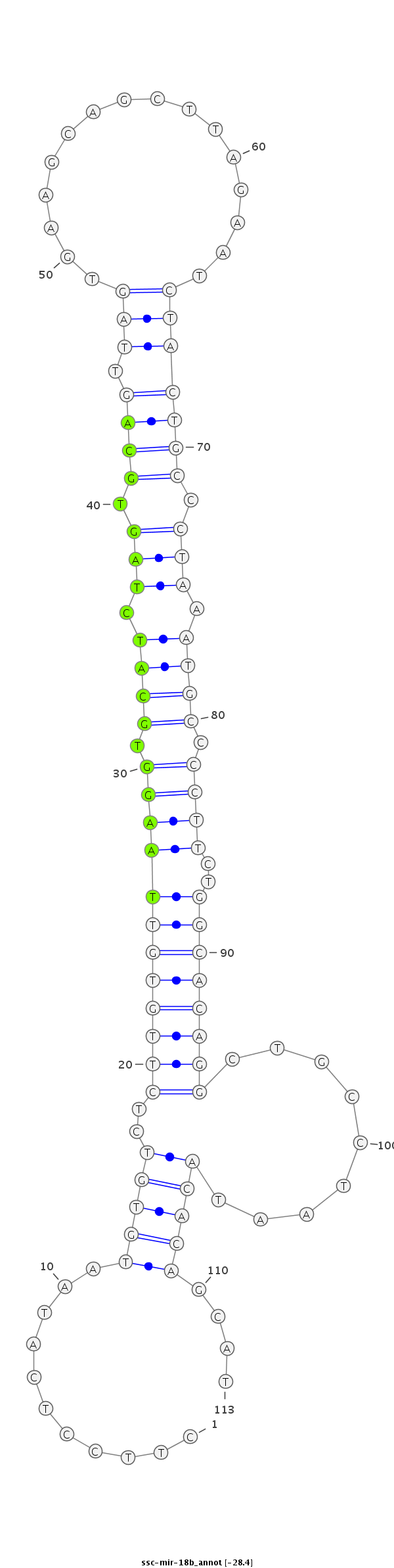
ID:ssc-mir-18b |
Coordinate:chrX:126200019-126200101 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -28.5 | -28.5 | -28.4 |
 |
 |
 |
| mature | star |
|
TTTGTGCATAATTAAGCTGTTGGTGCTTTTGAGCTCTTCCTCATAATGTGTCTCTTGTGTTAAGGTGCATCTAGTGCAGTTAGTGAAGCAGCTTAGAATCTACTGCCCTAAATGCCCCTTCTGGCACAGGCTGCCTAATACACAGCATGGAAAAATATGCCTTGATTAGTCGCTTGCATAGGA
***********************************...........(((((..((((((((((((.((((.(((.(((((.((................))))))).))).)))).)))..))))))))).........)))))....*********************************** |
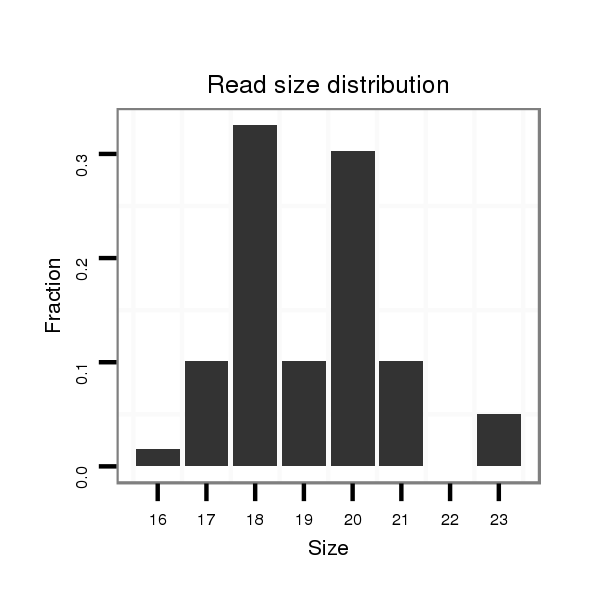
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR066810 testes |
SRR139075 testes |
SRR1274763 ovary |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TAAGGTGCATCTAGTGCA......................................................................................................... | 18 | 0 | 2 | 6.50 | 13 | 8 | 5 | 0 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGCAGT....................................................................................................... | 20 | 0 | 1 | 6.00 | 6 | 3 | 2 | 0 | 1 | 0 |
| ............................................................TAAGGTGCATCTAGTGCAG........................................................................................................ | 19 | 0 | 2 | 2.00 | 4 | 1 | 1 | 2 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGC.......................................................................................................... | 17 | 0 | 2 | 2.00 | 4 | 3 | 0 | 1 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGCAGTT...................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGCAGTTAG.................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGTAGTTAG.................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................ACTGCCCTAAATGCTCCTTCTT............................................................ | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGCAGGT...................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................GATTAAGGTGCATCTAGTGCA......................................................................................................... | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTG........................................................................................................... | 16 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TAAGGGGCATCTAGTGC.......................................................................................................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TAAGGTGCATCTAGTGAAA........................................................................................................ | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
AAACACGTATTAATTCGACAACCACGAAAACTCGAGAAGGAGTATTACACAGAGAACACAATTCCACGTAGATCACGTCAATCACTTCGTCGAATCTTAGATGACGGGATTTACGGGGAAGACCGTGTCCGACGGATTATGTGTCGTACCTTTTTATACGGAACTAATCAGCGAACGTATCCT
***********************************...........(((((..((((((((((((.((((.(((.(((((.((................))))))).))).)))).)))..))))))))).........)))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:45 AM