
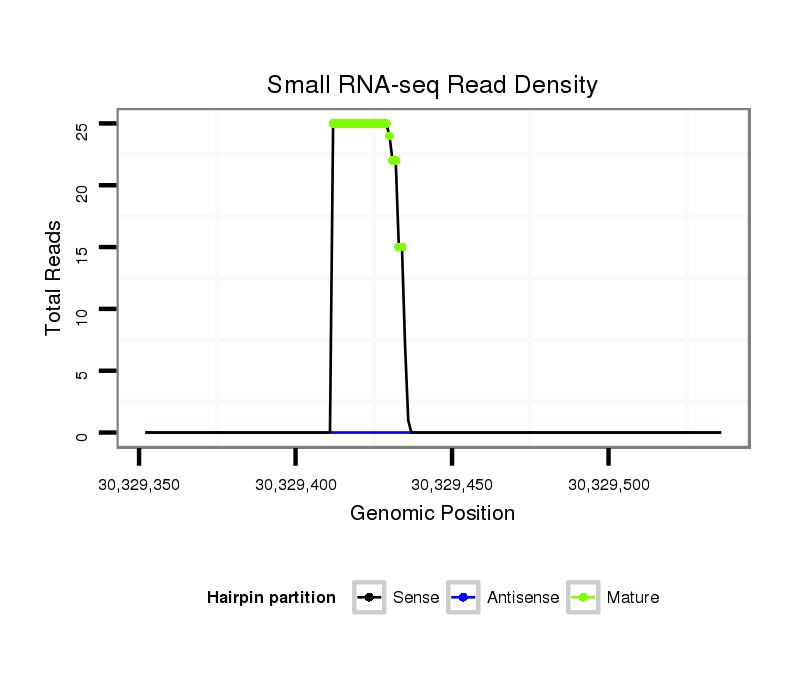
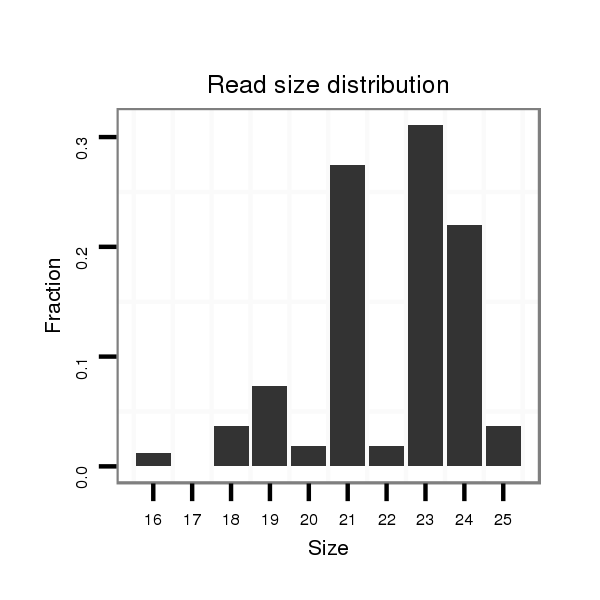
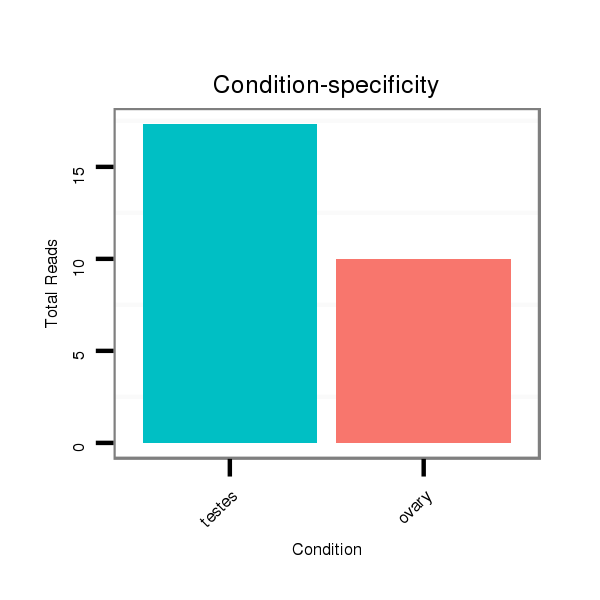
ID:ssc-mir-138 |
Coordinate:chr13:30329402-30329486 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -52.3 | -51.7 | -51.5 |
 |
 |
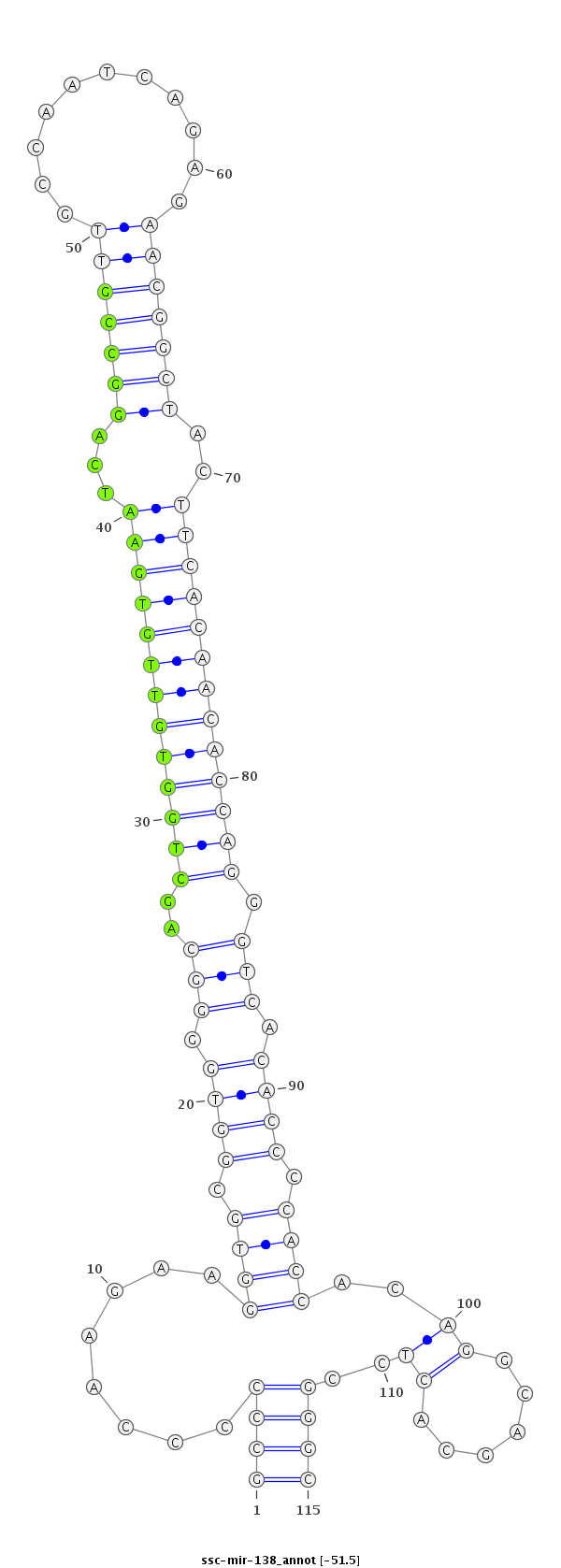 |
| mature | star |
|
AGGAAGGGACACCTCGACTGTCGGCTGCCCCAGAGGCCCCCCAAGAAGGTGCGGTGGGGCAGCTGGTGTTGTGAATCAGGCCGTTGCCAATCAGAGAACGGCTACTTCACAACACCAGGGTCACACCCCACCACAGGCAGCACTCCGGGCACCAGCCCCCATCGTGCTTCCACCTCAGCCATCGG
***********************************((((........((((((((((((...(((((((((((((...(((((((...........)))))))..))))))))))))).......))))))).......)))))..))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR139075 testes |
SRR066810 testes |
SRR1274764 testes |
SRR139076 testes |
SRR1274763 ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................AGCTGGTGTTGTGAATCAGGCCG...................................................................................................... | 23 | 0 | 2 | 8.50 | 17 | 7 | 7 | 1 | 1 | 0 | 1 |
| ............................................................AGCTGGTGTTGTGAATCAGGC........................................................................................................ | 21 | 0 | 2 | 7.50 | 15 | 3 | 5 | 6 | 0 | 1 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGT..................................................................................................... | 24 | 0 | 1 | 6.00 | 6 | 1 | 4 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCA....................................................................................................... | 22 | 1 | 2 | 2.50 | 5 | 2 | 3 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAG.......................................................................................................... | 19 | 0 | 2 | 2.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................CCGGCTACTTCACAACACCAGGGT................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGTAA................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCA........................................................................................................... | 18 | 0 | 2 | 1.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................GCTACTTCACAACACCAGGGG................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................CAACAAGGTGCGGTGGGGC............................................................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCAT...................................................................................................... | 23 | 2 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGTT.................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................CGGCTACTTCACAACACCAGGC................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCAA...................................................................................................... | 23 | 2 | 4 | 0.50 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCA...................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGC..................................................................................................... | 24 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCAG...................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGG..................................................................................................... | 24 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCGATA................................................................................................... | 26 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGG......................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGA........................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCCT...................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAATCAGGCC....................................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGAAT............................................................................................................. | 16 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................AGCTGGTGTTGTGCATCAGGCAT...................................................................................................... | 23 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................GCTGGTGTTGTGAATCAGGCA....................................................................................................... | 21 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
TCCTTCCCTGTGGAGCTGACAGCCGACGGGGTCTCCGGGGGGTTCTTCCACGCCACCCCGTCGACCACAACACTTAGTCCGGCAACGGTTAGTCTCTTGCCGATGAAGTGTTGTGGTCCCAGTGTGGGGTGGTGTCCGTCGTGAGGCCCGTGGTCGGGGGTAGCACGAAGGTGGAGTCGGTAGCC
***********************************((((........((((((((((((...(((((((((((((...(((((((...........)))))))..))))))))))))).......))))))).......)))))..))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:26 AM