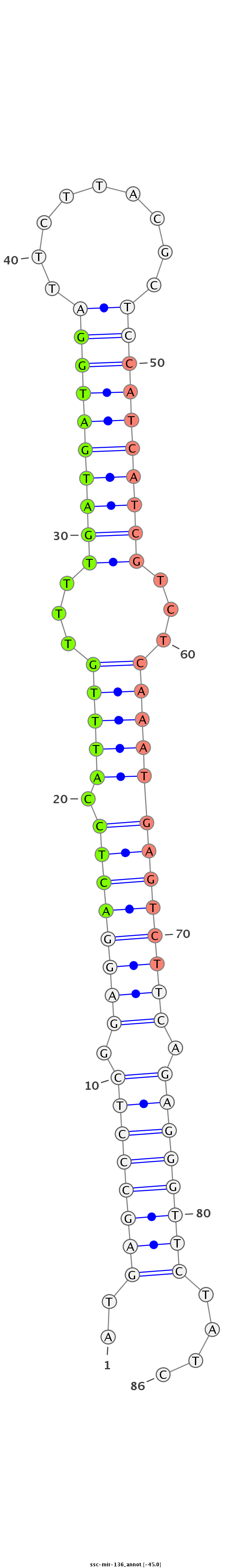
ID:ssc-mir-136 |
Coordinate:chr7:132079194-132079275 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
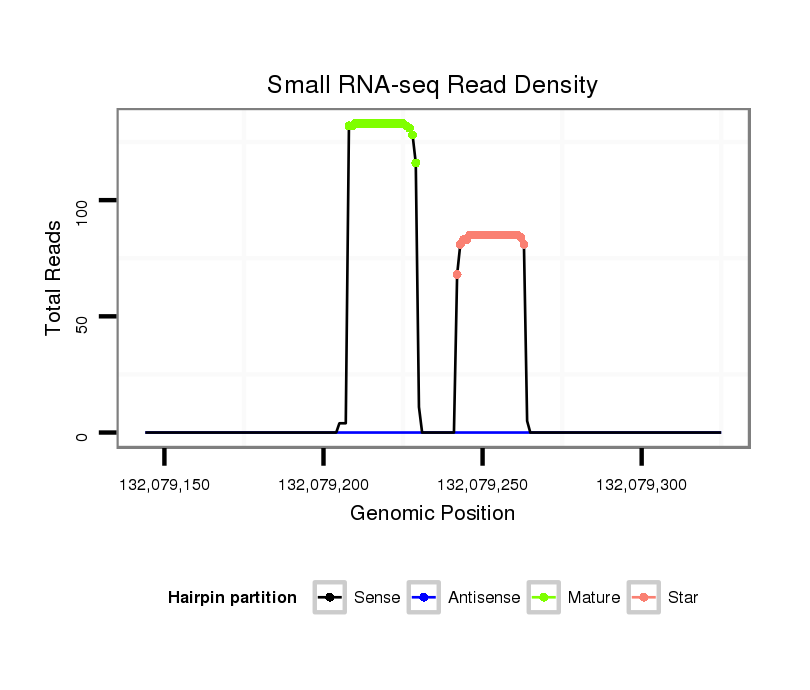
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -45.0 |
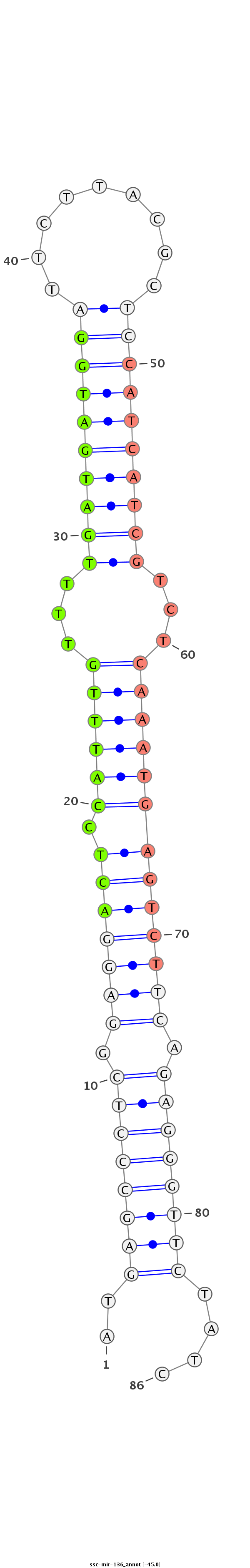 |
|
gccgccgcctgccgcGCTCCGCTGCCCGGCGTCTCCACGGTGGTGTTGGATGAGCCCTCGGAGGACTCCATTTGTTTTGATGATGGATTCTTACGCTCCATCATCGTCTCAAATGAGTCTTCAGAGGGTTCTATCATGTCGTCGGATGGAAAGGAGTGCCCTCTGAAGACGGGTAAGGTCGC
*************************************************..((((((((.((((((((.(((((...((((((((((.........))))))))))...))))))))))))).))))))))....*********************************************** |
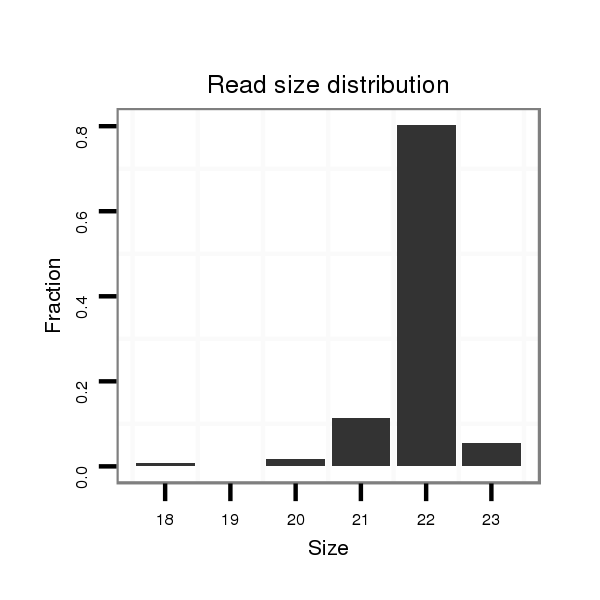
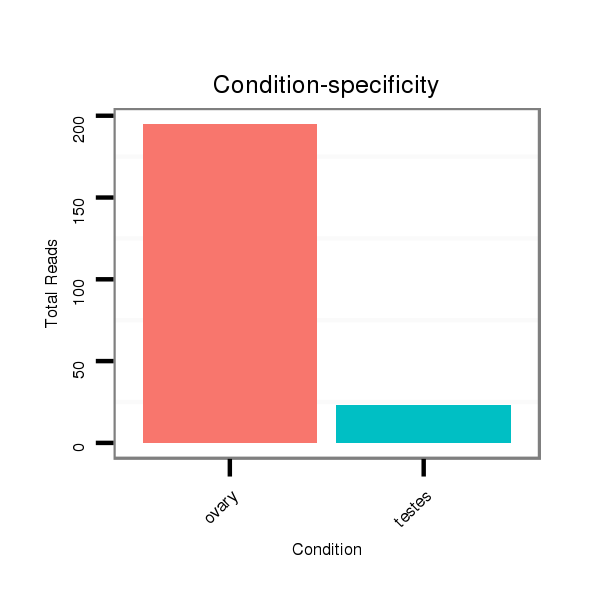
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR1274763 ovary |
SRR066810 testes |
SRR139075 testes |
SRR1274764 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................ACTCCATTTGTTTTGATGATGG................................................................................................ | 22 | 0 | 1 | 105.00 | 105 | 99 | 1 | 5 | 0 | 0 |
| ..................................................................................................CATCATCGTCTCAAATGAGTCT.............................................................. | 22 | 0 | 1 | 65.00 | 65 | 15 | 47 | 2 | 0 | 1 |
| ................................................................ACTCCATTTGTTTTGATGATG................................................................................................. | 21 | 0 | 1 | 12.00 | 12 | 3 | 6 | 2 | 0 | 1 |
| ................................................................ACTCCATTTGTTTTGATGATGGA............................................................................................... | 23 | 0 | 1 | 10.00 | 10 | 3 | 0 | 1 | 6 | 0 |
| ...................................................................................................ATCATCGTCTCAAATGAGTCT.............................................................. | 21 | 0 | 1 | 8.00 | 8 | 2 | 4 | 2 | 0 | 0 |
| ..................................................................................................CATCATTGTCTCAAATGAGTCT.............................................................. | 22 | 1 | 1 | 4.00 | 4 | 0 | 0 | 0 | 4 | 0 |
| ...................................................................................................ATCATCGTCTCAAATGAGTCTT............................................................. | 22 | 0 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................CACTCCATTTGTTTTGATGATGGA............................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................ATCGTCTCAAATGAGTCT.............................................................. | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................AGGACTCCATTTGTTTTGATGAT.................................................................................................. | 23 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................................CATCATCGTCTCAAATGAGTC............................................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................CATCGTCTCAAATGAGTCTTCT........................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................ACTCCATTTGTCTTGATGATGG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................ACTCCATTTGTTATGATGATGG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CATCATCGTCTCAAATGAGT................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................AGGACTCCATTTGTTTTGATGA................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................TCATCGTCTCAAATGAGTCT.............................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................ACTCCATTTGTTTTGATGAT.................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................ACTCCATTTGTTTTAATGATGG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CATCATCGTCTCAAATGAGTCTA............................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................TCATCGTCTCAAATGAGTCTT............................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................ATCATCGTCTCAAATGAGTC............................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................TCTCCATTTGTTTTGATGATGG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................AGGACTCCATTTGTTTTGATG.................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................ACTCCATTTGTTTTGATGATGGAA.............................................................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................TATCATCGTCTCAAATGAGT................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................ATCATCGTCTCAAATGAGCCT.............................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CATCATCGTCTCAAATGAGTCAA............................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................TCCATTTGTTTTGATGATGGA............................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................ACTCCATTTATTTTGATGATGG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................ATTTGTTGTGATGATGG................................................................................................ | 17 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
gGGgGGgGGtgGGgGCGAGGCGACGGGCCGCAGAGGTGCCACCACAACCTACTCGGGAGCCTCCTGAGGTAAACAAAACTACTACCTAAGAATGCGAGGTAGTAGCAGAGTTTACTCAGAAGTCTCCCAAGATAGTACAGCAGCCTACCTTTCCTCACGGGAGACTTCTGCCCATTCCAGCG
***********************************************..((((((((.((((((((.(((((...((((((((((.........))))))))))...))))))))))))).))))))))....************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139075 testes |
SRR139076 testes |
|---|---|---|---|---|---|---|---|
| .........................................................................................................................................CAGCAGCCTACATTT.............................. | 15 | 1 | 11 | 0.27 | 3 | 2 | 1 |
| .........................................................................................................................................CAGCAGCCTACATTTG............................. | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 |
Generated: 09/01/2015 at 11:40 AM