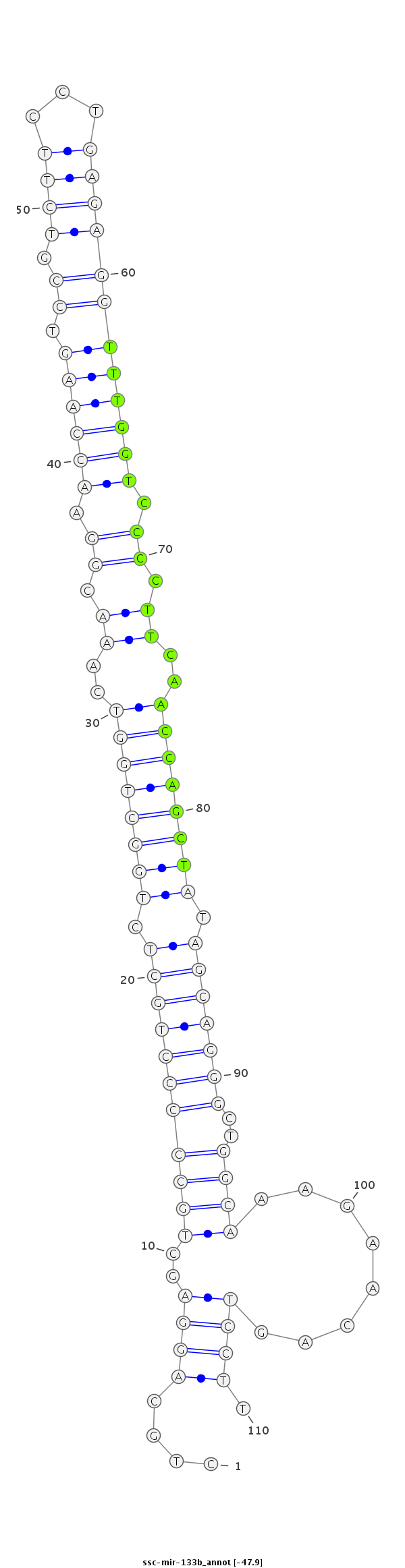
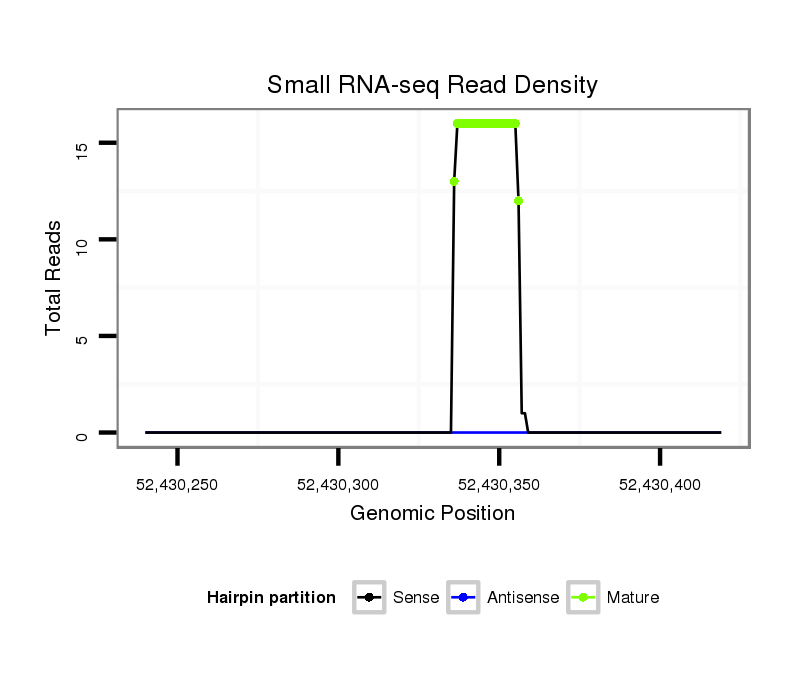
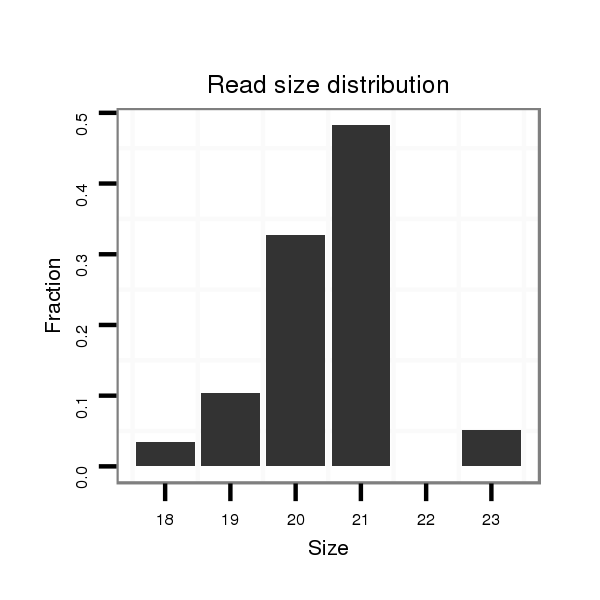
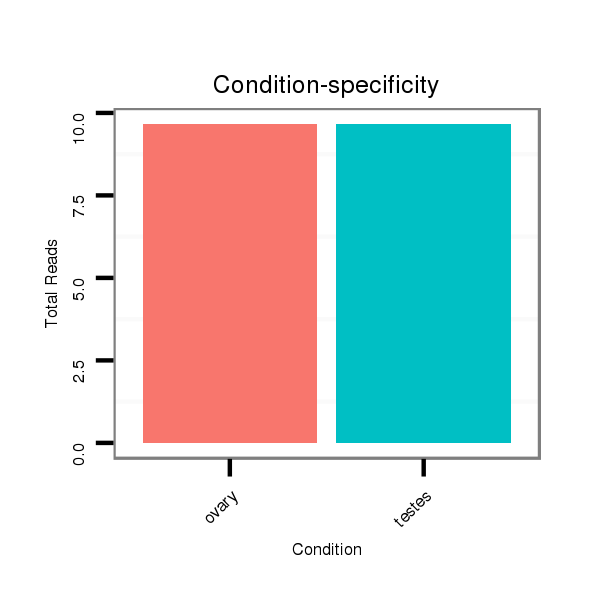
ID:ssc-mir-133b |
Coordinate:chr7:52430290-52430369 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -47.1 | -47.1 | -47.0 |
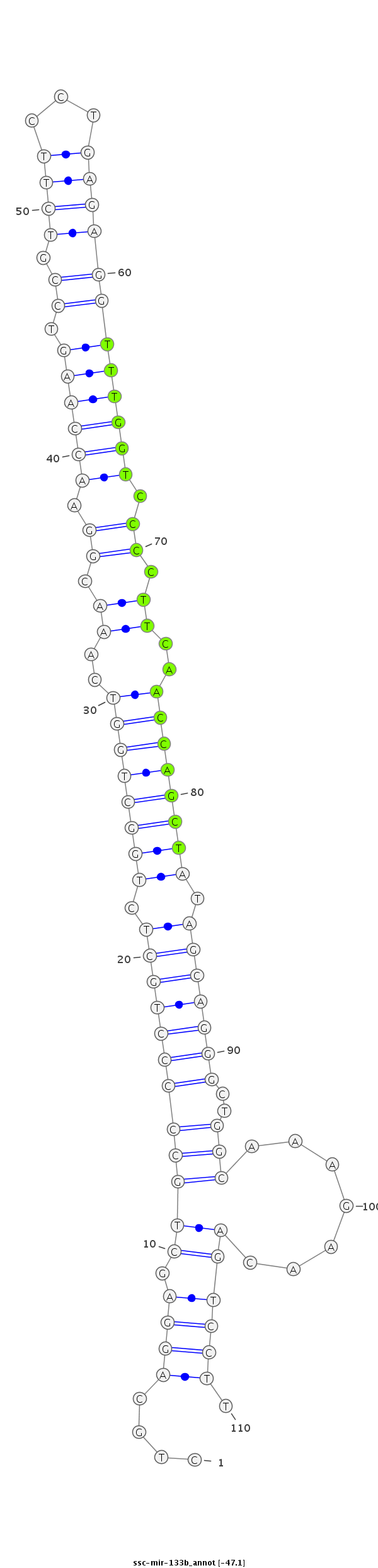 |
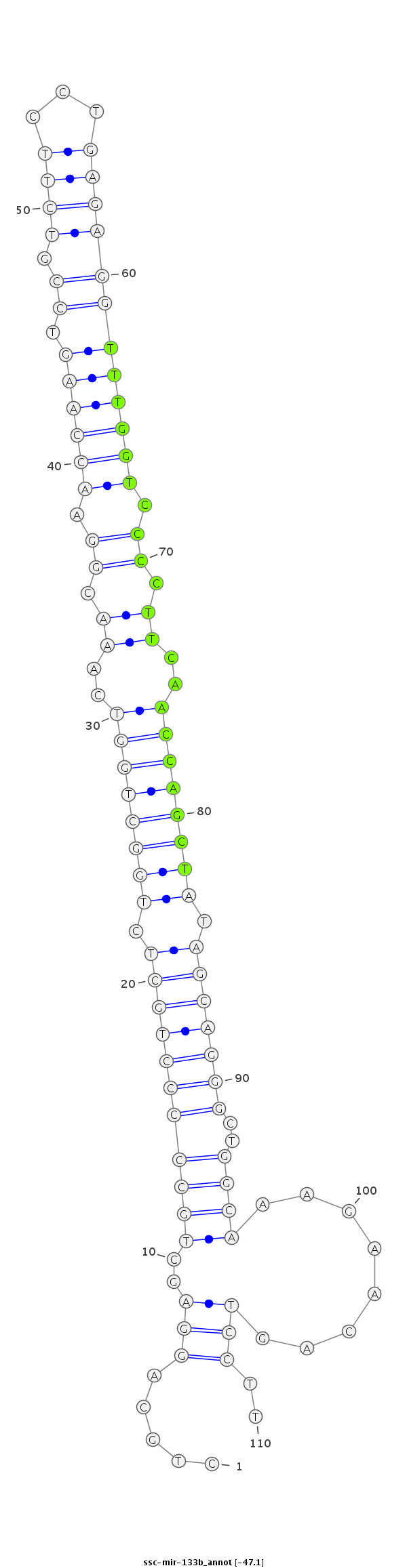 |
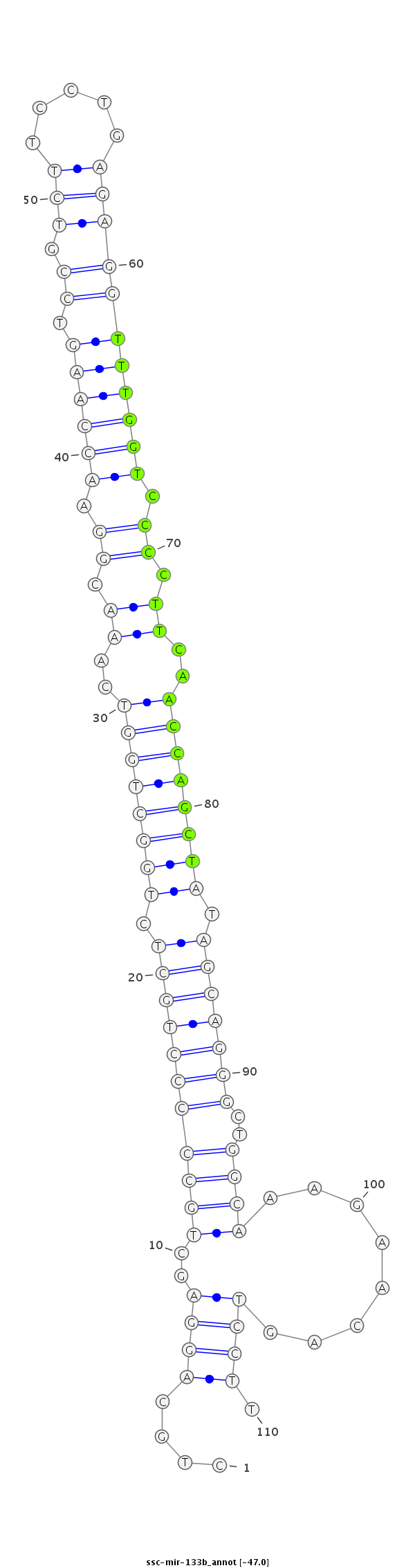 |
| mature | star |
|
CCAGGAATCCTCACCCTAGAGGCTGCAGTCACCTCCTGCAGGAGCTGCCCCCTGCTCTGGCTGGTCAAACGGAACCAAGTCCGTCTTCCTGAGAGGTTTGGTCCCCTTCAACCAGCTATAGCAGGGCTGGCAAAGAACAGTCCTTGGAGGAACAGAAGAGATTCCACTGTGGCTGAAATT
***********************************....((((..(((((((((((.((((((((..((.((.((((((.((.((((...)))))))))))).)).))..)))))))).)))))))..))))........)))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR139075 testes |
SRR1274763 ovary |
SRR139076 testes |
SRR066810 testes |
SRR1274764 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................TTTGGTCCCCTTCAACCAGCT............................................................... | 21 | 0 | 3 | 9.33 | 28 | 11 | 9 | 4 | 1 | 2 | 1 |
| ................................................................................................TTTGGTCCCCTTCAACCAGC................................................................ | 20 | 0 | 3 | 3.67 | 11 | 4 | 2 | 4 | 1 | 0 | 0 |
| .................................................................................................TTGGTCCCCTTCAACCAGCT............................................................... | 20 | 0 | 3 | 2.67 | 8 | 3 | 0 | 3 | 1 | 0 | 1 |
| .................................................................................................TTGGTCCCCTTCAACCAGC................................................................ | 19 | 0 | 3 | 1.33 | 4 | 0 | 0 | 0 | 0 | 3 | 1 |
| ................................................................................................TTTGGTCCCCTTCAACCAGCTAT............................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGGTCCCCTTCAACCAGCTAAA............................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................TTGGTCCCCTTCAACCAGCTTT............................................................. | 22 | 1 | 3 | 0.67 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................TTTGGTCCCCTTCAACCAGCTT.............................................................. | 22 | 1 | 3 | 0.67 | 2 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................TTTGGTCCCCTTCAACCAG................................................................. | 19 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................GGTCCCCTTCAACCAGCT............................................................... | 18 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................TGGTCCCCTTCAACCAGCTCT............................................................. | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGGTCCCCTTCAACCAGCTT.............................................................. | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................TTTGGTCCCTTTCAACCAGCTT.............................................................. | 22 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GGTCCTTAGGAGTGGGATCTCCGACGTCAGTGGAGGACGTCCTCGACGGGGGACGAGACCGACCAGTTTGCCTTGGTTCAGGCAGAAGGACTCTCCAAACCAGGGGAAGTTGGTCGATATCGTCCCGACCGTTTCTTGTCAGGAACCTCCTTGTCTTCTCTAAGGTGACACCGACTTTAA
***********************************....((((..(((((((((((.((((((((..((.((.((((((.((.((((...)))))))))))).)).))..)))))))).)))))))..))))........)))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:40 AM