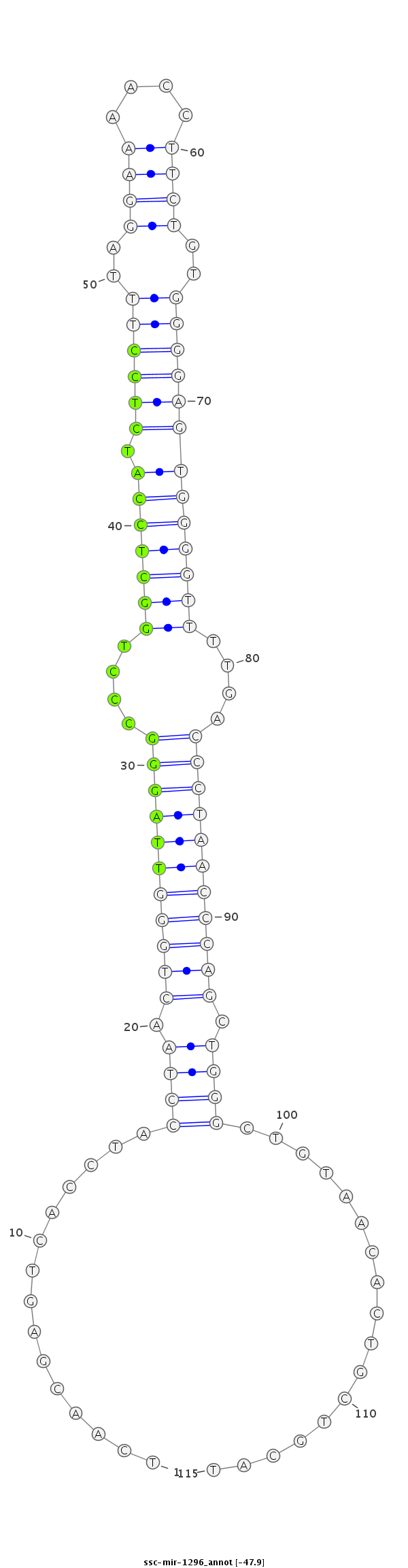
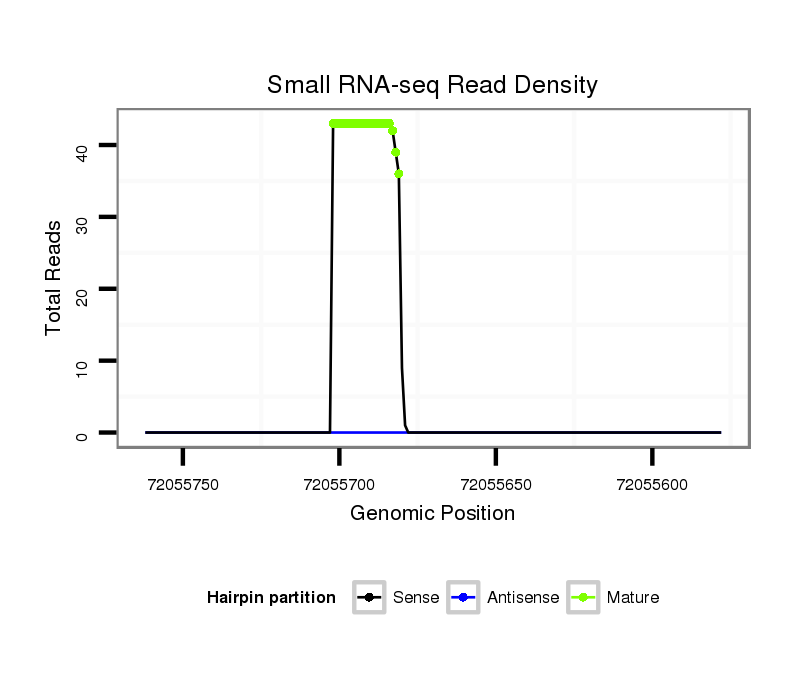
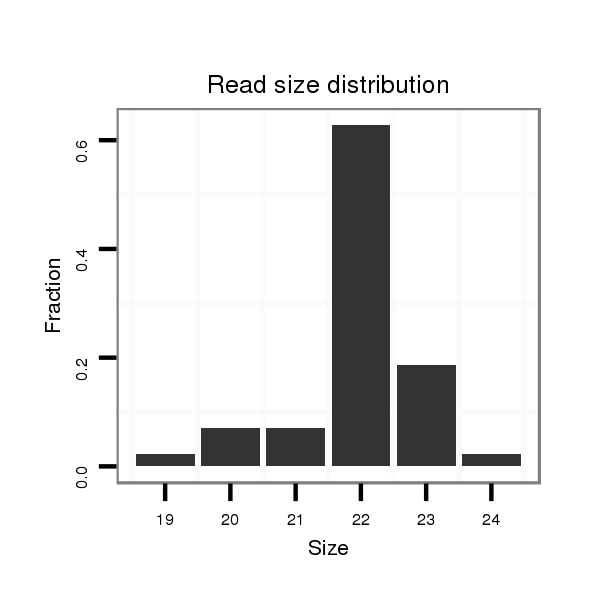
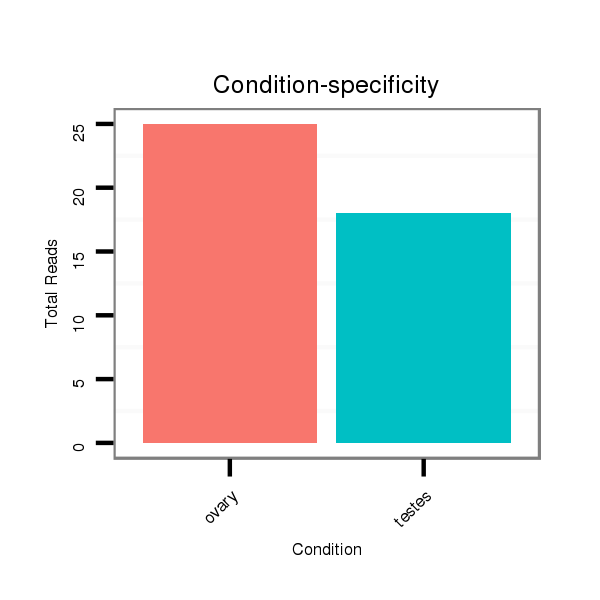
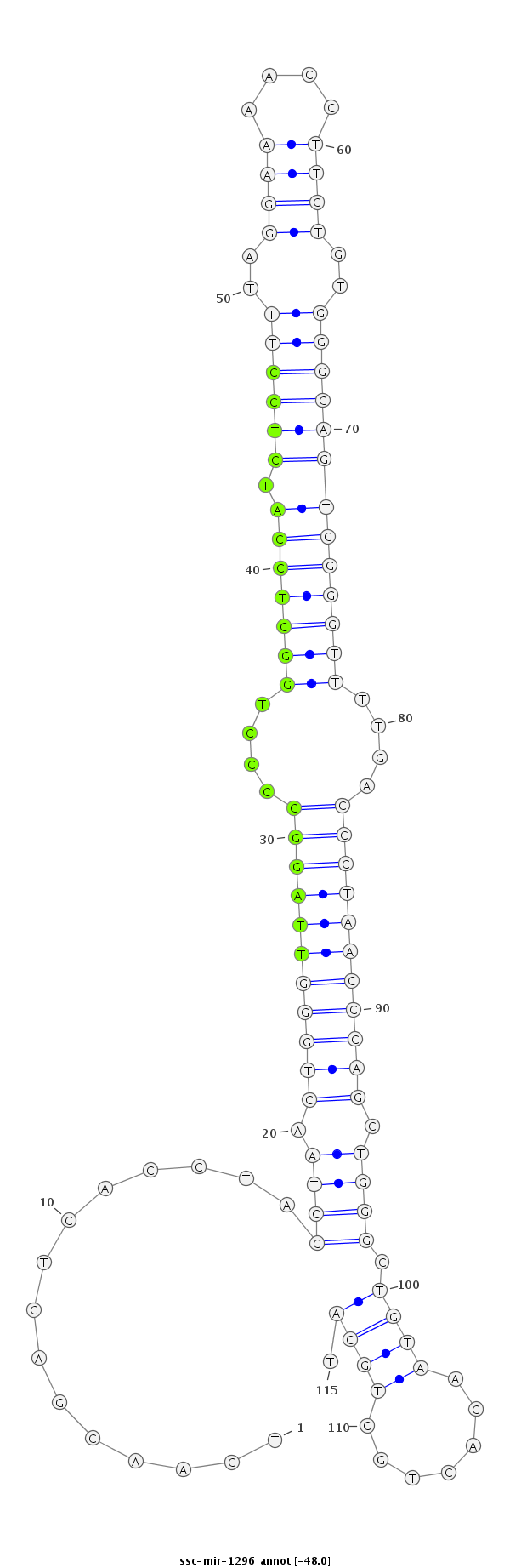
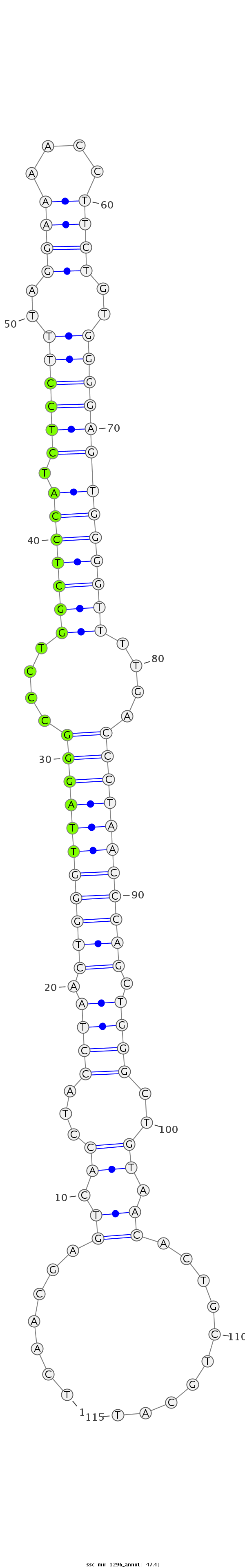
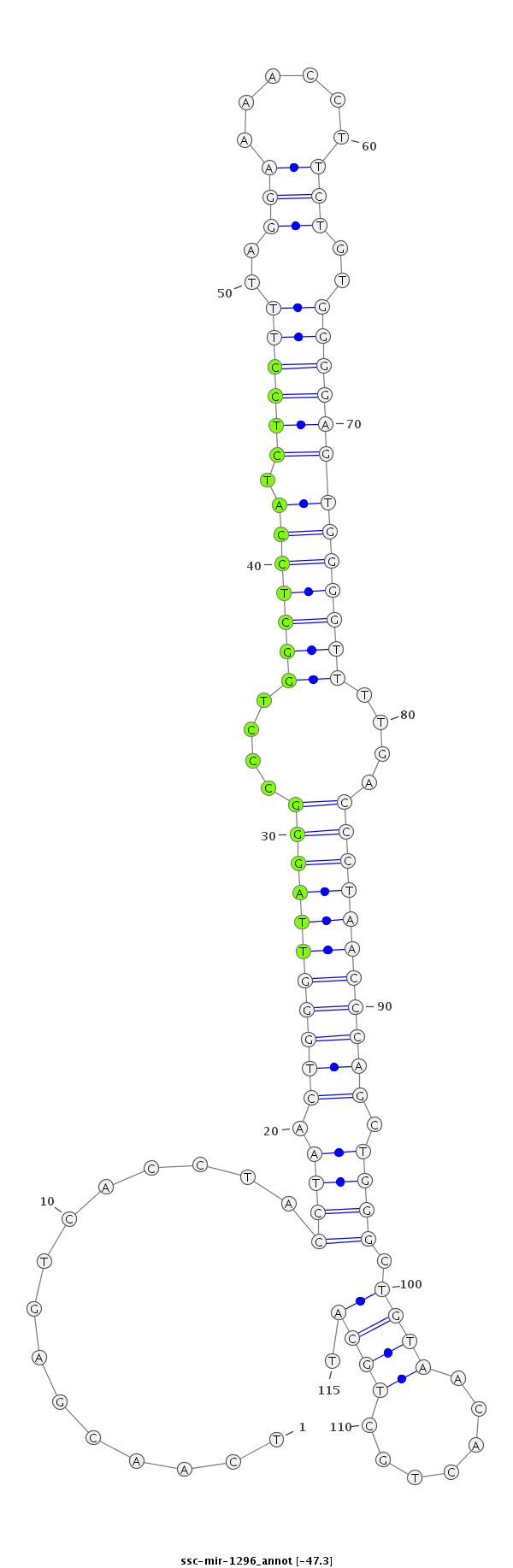
ID:ssc-mir-1296 |
Coordinate:chr14:72055628-72055712 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -48.0 | -47.4 | -47.3 |
 |
 |
 |
|
ACCAGGGAAAGAAAGTGTTGCATTCATATTCGAAATCAACGAGTCACCTACCTAACTGGGTTAGGGCCCTGGCTCCATCTCCTTTAGGAAAACCTTCTGTGGGGAGTGGGGTTTTGACCCTAACCCAGCTGGGCTGTAACACTGCTGCATGTTCTAAGGGGCTGAGTTTTCTACTACTTTTCTGC
***********************************...............((((.(((((((((((....(((((((.((((((..((((....))))..)))))))))))))....))))))))))).)))).................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR066809 ovary |
SRR066810 testes |
SRR1274763 ovary |
SRR1274764 testes |
SRR139076 testes |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TTAGGGCCCTGGCTCCATCTCC....................................................................................................... | 22 | 0 | 1 | 27.00 | 27 | 11 | 8 | 2 | 6 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCT...................................................................................................... | 23 | 0 | 1 | 8.00 | 8 | 2 | 0 | 4 | 2 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCT....................................................................................................... | 22 | 1 | 1 | 3.00 | 3 | 1 | 1 | 0 | 1 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCAAT.................................................................................................... | 25 | 2 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 |
| ............................................................TTAGGGCCCTGGCTCCATCTC........................................................................................................ | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCATT.................................................................................................... | 25 | 1 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCAT..................................................................................................... | 24 | 1 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCT......................................................................................................... | 20 | 0 | 1 | 3.00 | 3 | 0 | 2 | 1 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCA...................................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCA....................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCA......................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATC.......................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCAA...................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCTATA................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCCTT..................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTAGGGCCCTGGCTCCATCTCAGA..................................................................................................... | 24 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................CTGGGCTGTAACACTGG........................................ | 17 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TGGTCCCTTTCTTTCACAACGTAAGTATAAGCTTTAGTTGCTCAGTGGATGGATTGACCCAATCCCGGGACCGAGGTAGAGGAAATCCTTTTGGAAGACACCCCTCACCCCAAAACTGGGATTGGGTCGACCCGACATTGTGACGACGTACAAGATTCCCCGACTCAAAAGATGATGAAAAGACG
***********************************...............((((.(((((((((((....(((((((.((((((..((((....))))..)))))))))))))....))))))))))).)))).................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:27 AM