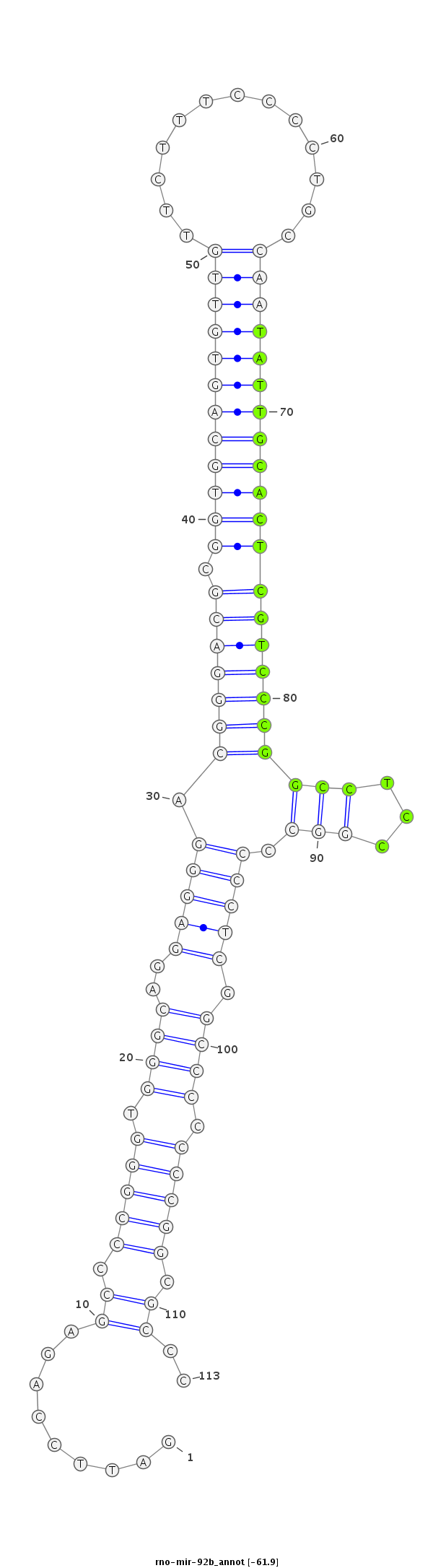
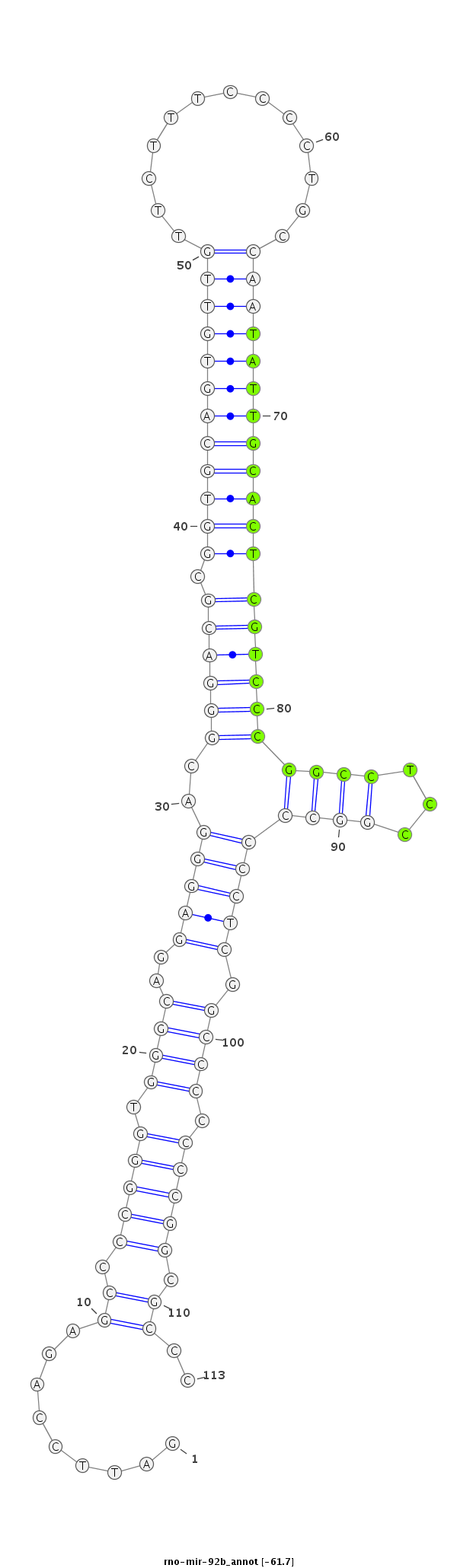
ID:rno-mir-92b |
Coordinate:chr2:188541034-188541116 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -61.9 | -61.7 |
 |
 |
|
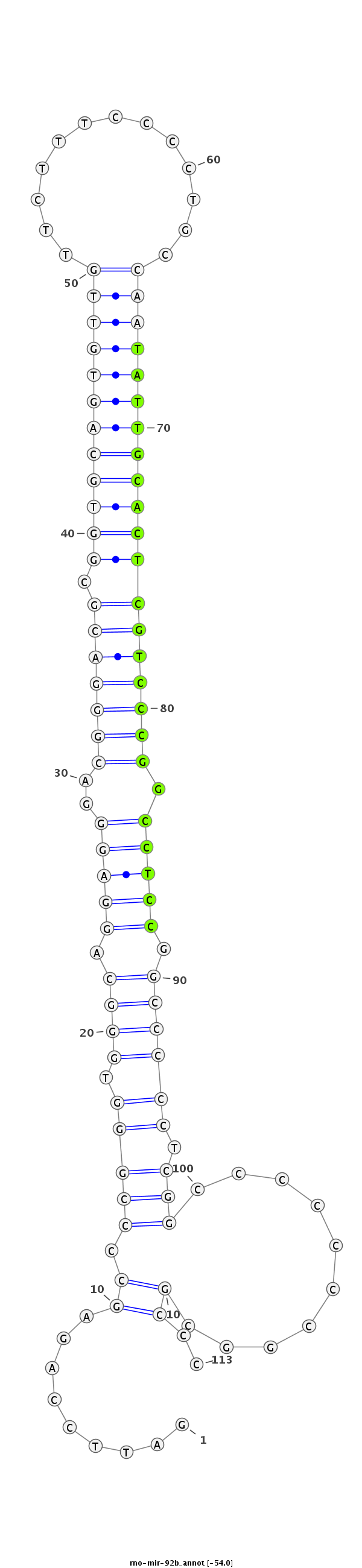
CCCTGGGCTCCAGCGACCCAGGAGGTAGGGAGCGGGATTCCAGAGCCCCGGGTGGGCAGGAGGGACGGGACGCGGTGCAGTGTTGTTCTTTCCCCTGCCAATATTGCACTCGTCCCGGCCTCCGGCCCCCTCGGCCCCCCCGGCGCCCCCTCACTCAGAGTGGGGCAGCCCCAGCCACCGGGC
***********************************.........((.(((((.((((.(((((..(((((((.((((((((((((.............))))))))))))))))))).))))).)))))).)))..........))..*********************************** |
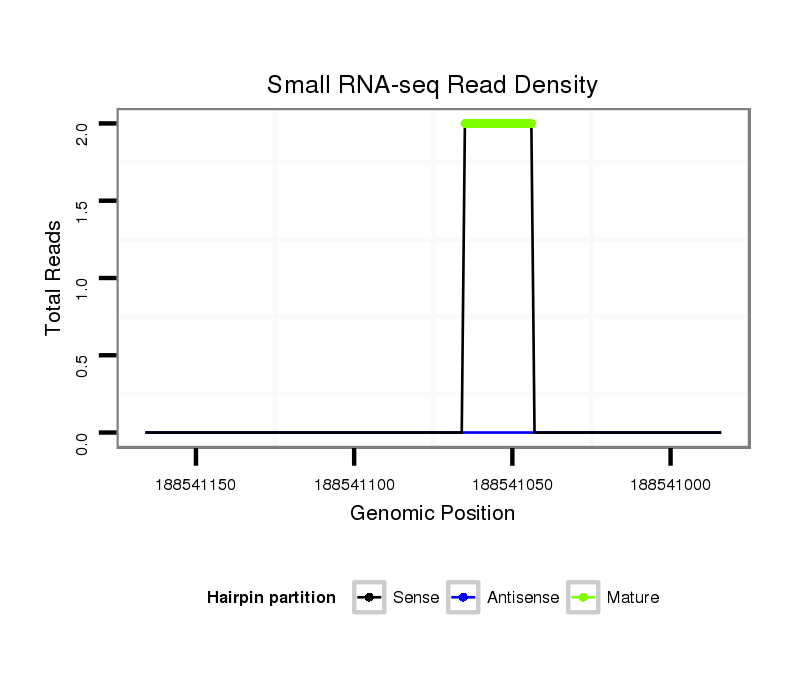
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
GSM1528817 Adult_testes |
GSM1528818 Adult_testes_Mili_IP |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TATTGCACTCGTCCCGGCCTCAA........................................................... | 23 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ....................................................................................................ATATTGCACTTGTCCCGGCCT.............................................................. | 21 | 1 | 3 | 2.00 | 6 | 0 | 6 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCCT........................................................... | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCC............................................................ | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .....................................................................................................TATTGCCCTCGTCCCGGCCTCAT........................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................................TTGCACTCGTCCCGGCCTCCAA.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCCTTA......................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................................................................................ATTGCACTCGTCCCGGCCTCCT........................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCTAT.......................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................................................................................ATTGCACTCGTCCCGGCCTCCTAT......................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCCTAT......................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCAAG.......................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCATA.......................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................................GTATATTGCACTTGTCCCGGCCT.............................................................. | 23 | 3 | 3 | 1.00 | 3 | 2 | 0 | 1 |
| ....................................................................................................ATATTGCACTTGTCCCGGC................................................................ | 19 | 1 | 3 | 0.67 | 2 | 0 | 2 | 0 |
| ......................................................................................................ATTGCACTCGTCCCGGCCTCCTTT......................................................... | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .....................................................................................................AATTGCACTAGTCCCGGCCTGCG........................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTCTT........................................................... | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTTAA........................................................... | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTCGTCCCGGCCTATA........................................................... | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................ATATTGCACTTGTCCCGGCCTCT............................................................ | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................ATATTGCACTTGTCCCGGCC............................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ....................................................................................................ATATTGCACTTGTACCGGCCT.............................................................. | 21 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .....................................................................................................TATTGCACTTGTCCCGGCCTAC............................................................ | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................ATATTGCACTTGTCCCGGCCTGC............................................................ | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................ATATTCCACTTGTCCCGGCCT.............................................................. | 21 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ....................................................................................................ATATTGCACTTGTCCCGG................................................................. | 18 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 |
|
GGGACCCGAGGTCGCTGGGTCCTCCATCCCTCGCCCTAAGGTCTCGGGGCCCACCCGTCCTCCCTGCCCTGCGCCACGTCACAACAAGAAAGGGGACGGTTATAACGTGAGCAGGGCCGGAGGCCGGGGGAGCCGGGGGGGCCGCGGGGGAGTGAGTCTCACCCCGTCGGGGTCGGTGGCCCG
***********************************.........((.(((((.((((.(((((..(((((((.((((((((((((.............))))))))))))))))))).))))).)))))).)))..........))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
|---|---|---|---|---|---|---|
| .....................................................................................................................GGGAGGCCGAGGGAGCCGGGGC............................................ | 22 | 3 | 20 | 0.05 | 1 | 1 |
Generated: 04/30/2015 at 05:42 PM