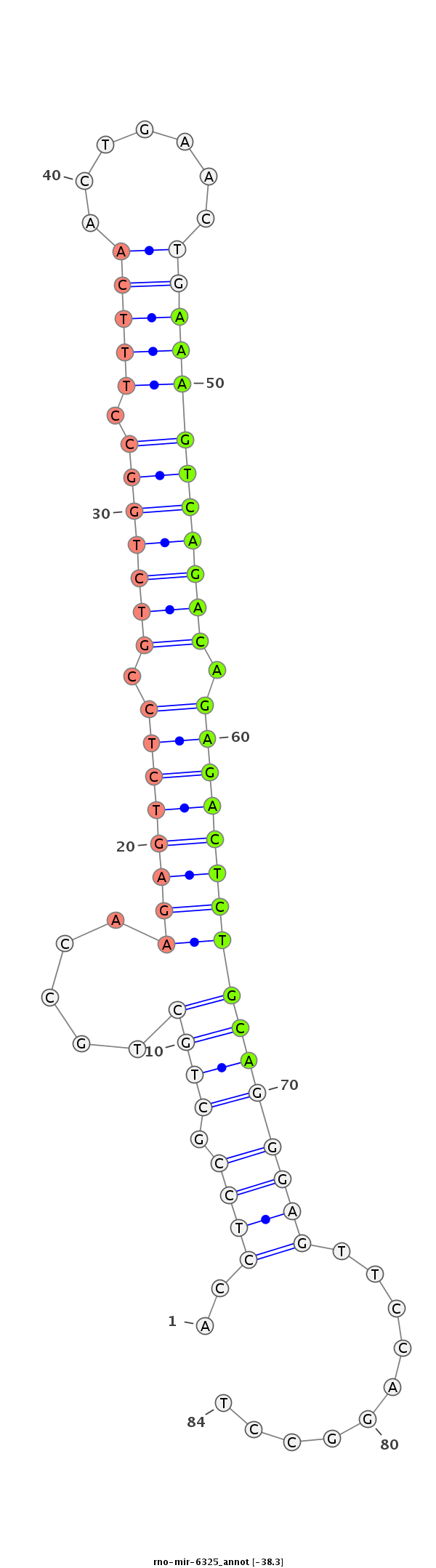
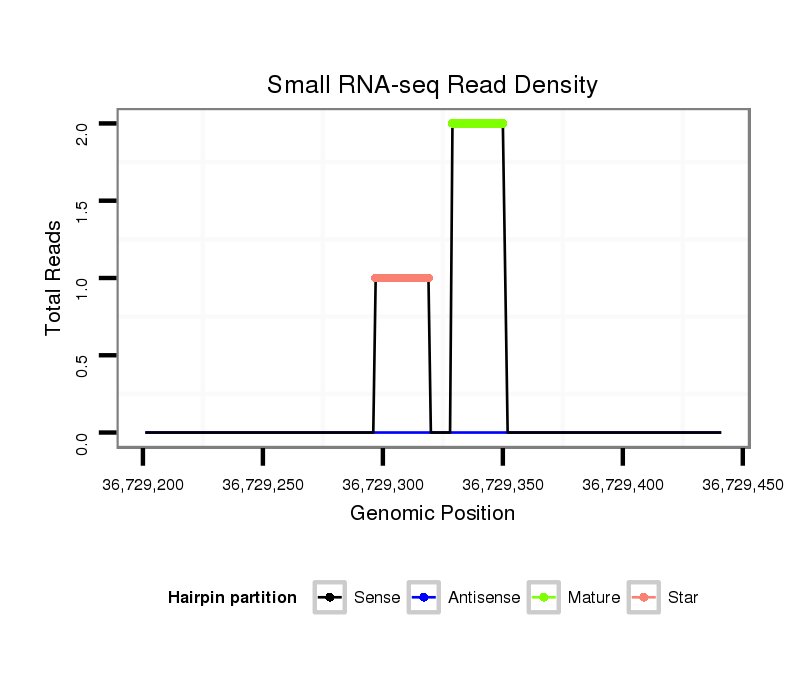
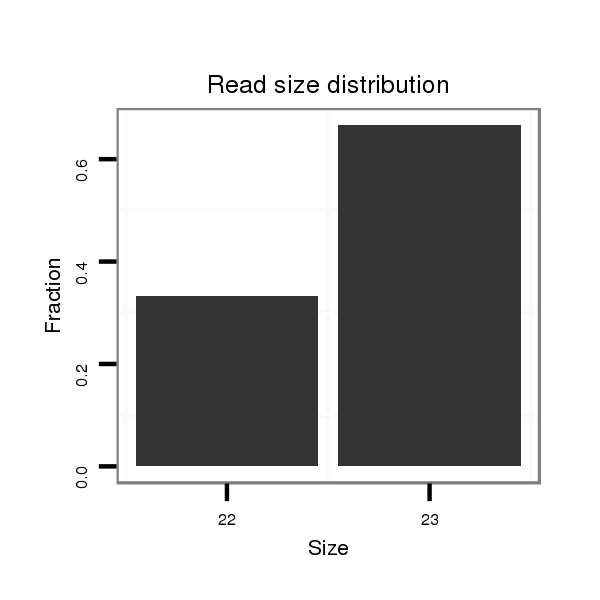
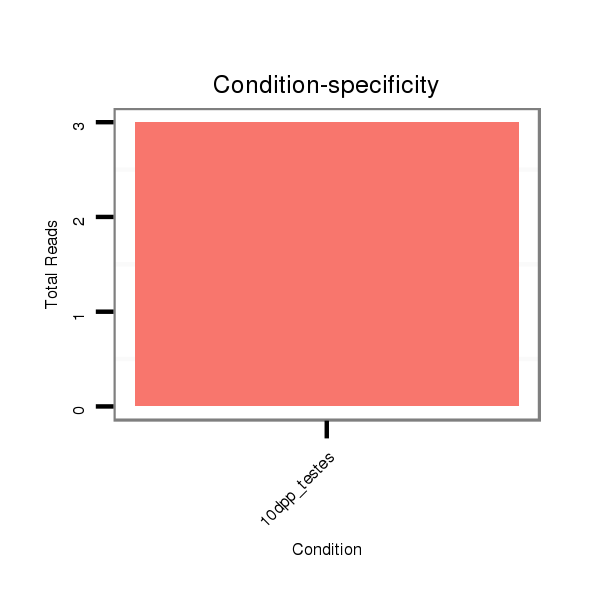
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCAA.......................................................................................... |
23 |
1 |
1 |
4.00 |
4 |
4 |
0 |
| .................................................................................................................................AAGTCAGACAGAGACTCTGCAA.......................................................................................... |
22 |
1 |
1 |
3.00 |
3 |
3 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGTAA.......................................................................................... |
23 |
2 |
2 |
1.00 |
2 |
2 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCAGT......................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGCCTCTGCAGA......................................................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................................................................................AAGTCAGACAGAGACTCTGCAGA......................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCAGA......................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCA........................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGTTG.......................................................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................AAGAGTCTCCGTCTGGCCTTTCA.......................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGA............................................................................................ |
21 |
1 |
1 |
1.00 |
1 |
0 |
1 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCAAT......................................................................................... |
24 |
2 |
2 |
1.00 |
2 |
2 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGT............................................................................................ |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCAG.......................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................AAGAGTCTCCGTCTGGCCTTTAA.......................................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGCTAT......................................................................................... |
24 |
3 |
5 |
0.20 |
1 |
1 |
0 |
| ................................................................................................................................AAAGTCAGACAGAGACTCTGTTA.......................................................................................... |
23 |
3 |
13 |
0.08 |
1 |
1 |
0 |