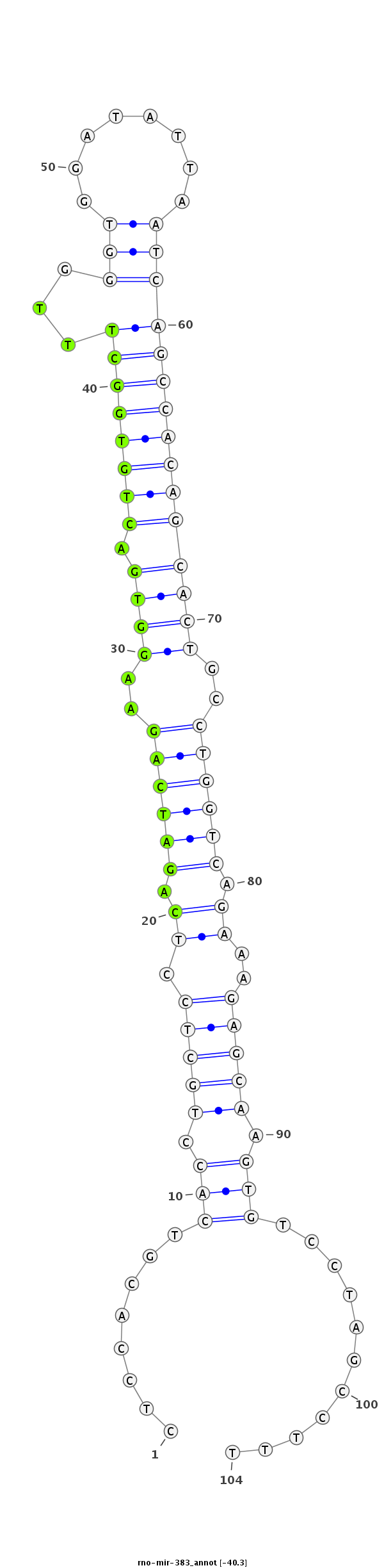
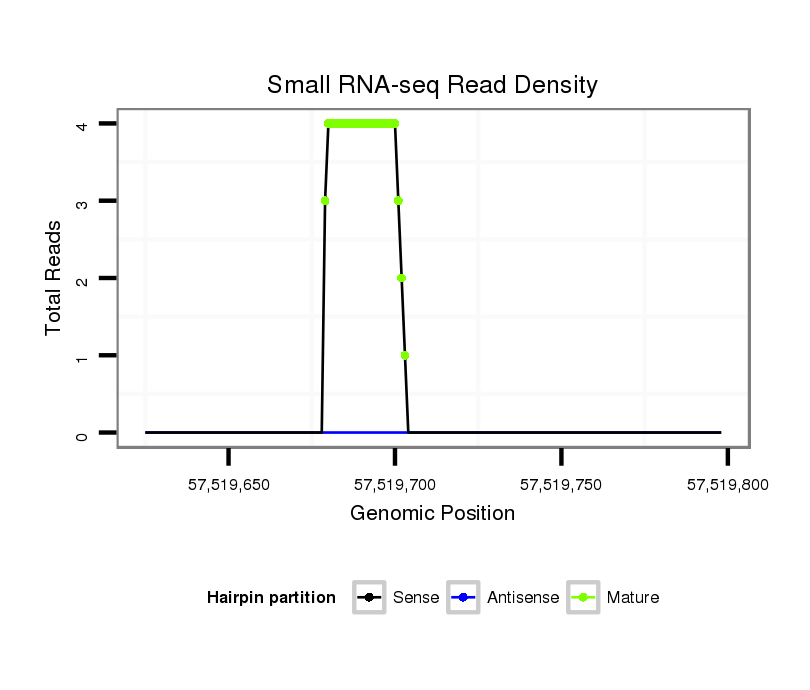
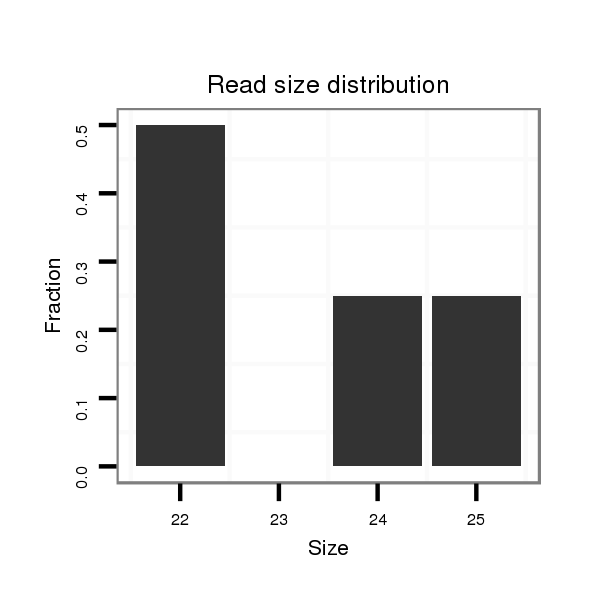
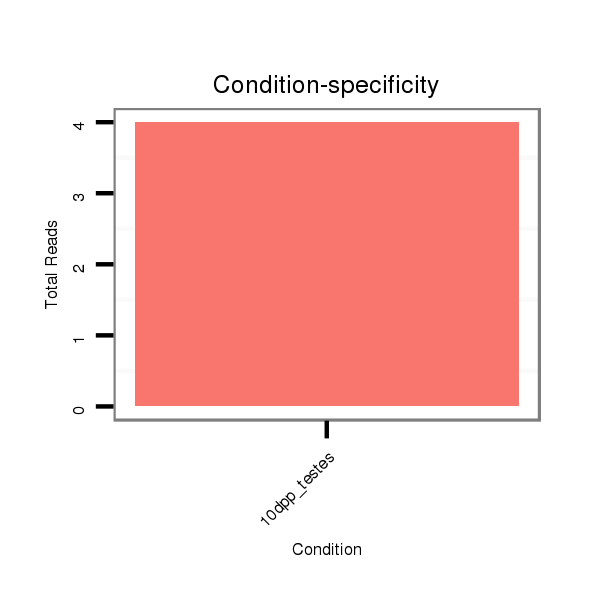
ID:rno-mir-383 |
Coordinate:chr16:57519675-57519748 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
TTATGCTTACGTAATTTATCAAGAGCCTCCAGAAACTCCACGTCACCTGCTCCTCAGATCAGAAGGTGACTGTGGCTTTGGGTGGATATTAATCAGCCACAGCACTGCCTGGTCAGAAAGAGCAAGTGTCCTAGCCTTTTACCTCATGACACCTTGTGGGTCCATCCAGCATAG
***********************************........(((.(((((.((.((((((..((((.((((((((...(((........)))))))))))))))..)))))).))..))))).)))...........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
GSM1528818 Adult_testes_Mili_IP |
|---|---|---|---|---|---|---|---|
| .......................................................AGATCAGAAGGTGACTGTGGCTA................................................................................................ | 23 | 1 | 1 | 4.00 | 4 | 4 | 0 |
| .......................................................AGATCAGAAGGTGACTGTGGCTAT............................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGCAAT............................................................................................... | 25 | 2 | 1 | 2.00 | 2 | 2 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGCTTT............................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGCTT................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGCTA................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| .......................................................AGATCAGAAGGTGACTGTGGCT................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................CAGCTCAGAAGGTGACTGTGGCAT................................................................................................ | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGCTTA............................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 |
| ......................................................CAGATCAGAAGGTGACTGTGGC.................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .......................................................AGATCAGAAGGTGACTGTGGCTTTA.............................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................CAGATCAGAAGGTGACTGTGGTA................................................................................................. | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| .................................................................................................CACAGCACTGCCTGGTCAGAATTT..................................................... | 24 | 3 | 4 | 0.25 | 1 | 1 | 0 |
|
AATACGAATGCATTAAATAGTTCTCGGAGGTCTTTGAGGTGCAGTGGACGAGGAGTCTAGTCTTCCACTGACACCGAAACCCACCTATAATTAGTCGGTGTCGTGACGGACCAGTCTTTCTCGTTCACAGGATCGGAAAATGGAGTACTGTGGAACACCCAGGTAGGTCGTATC
***********************************........(((.(((((.((.((((((..((((.((((((((...(((........)))))))))))))))..)))))).))..))))).)))...........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 04/30/2015 at 05:36 PM