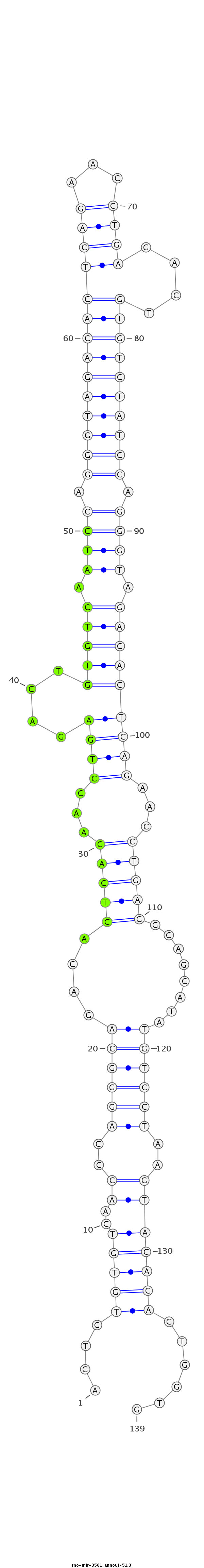
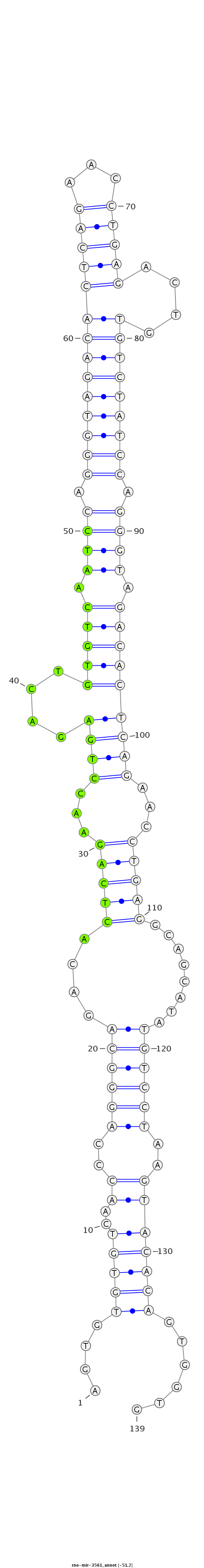
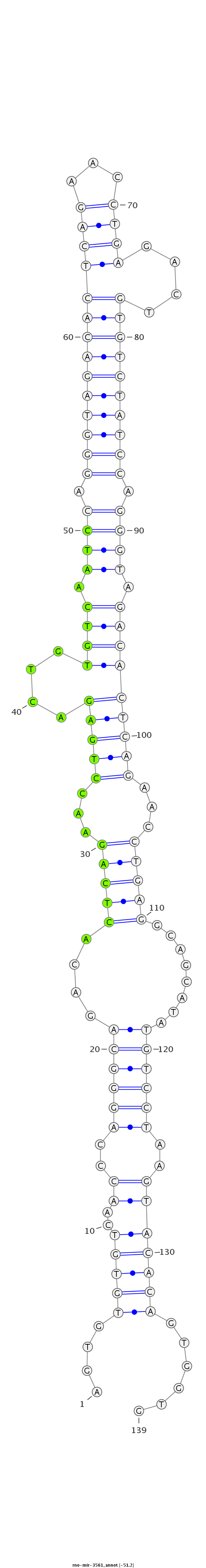
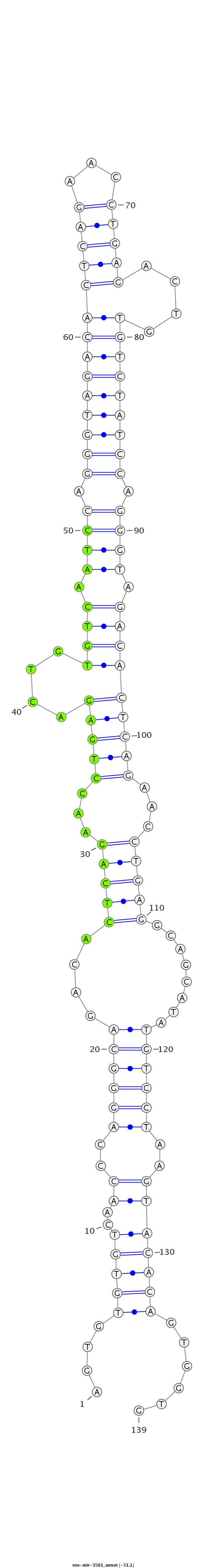
ID:rno-mir-3561 |
Coordinate:chr7:26427720-26427828 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -51.2 | -51.2 | -51.1 |
 |
 |
 |
|
GTGTCAACCCAGGGCAGACACTCAGAACCTGTGACAGTGTGTGTCAACCCAGGGCAGACACTCAGAACCTGAGACTGTGTCAATCCAGGGTAGACACTCAGAACCTGAGACTGTGTCTATCCAGGGTAGACACTCAGAACCTGAGGCAGCATATGTCCTAAGTACACAGTGGTGTCTCACAAGCCAGTGAAAACAGATAGAAGCCAAAT
***********************************....(((((..((..((((((....(((((...((((....(((((.((((.((((((((((((((...))))....)))))))))).)))).)))))))))...)))))........))))))..)))))))......*********************************** |
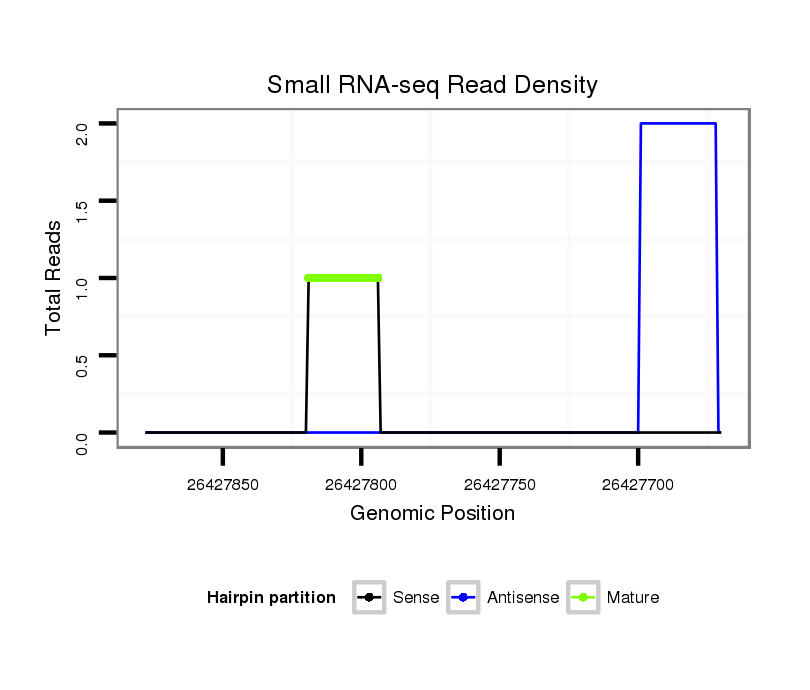
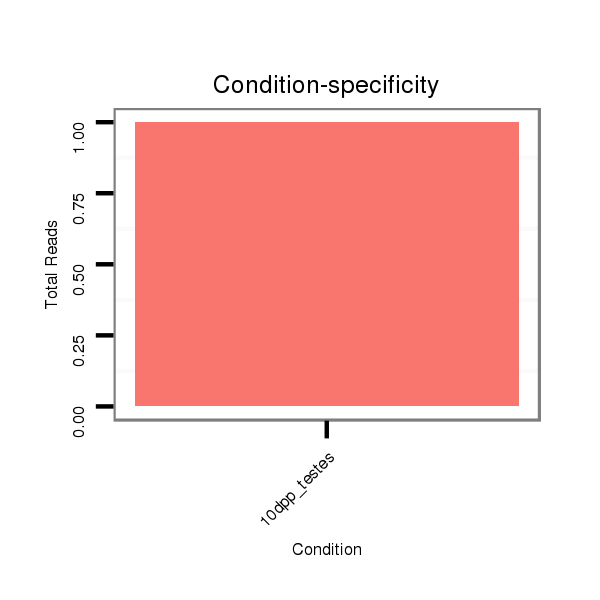
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
|---|---|---|---|---|---|---|
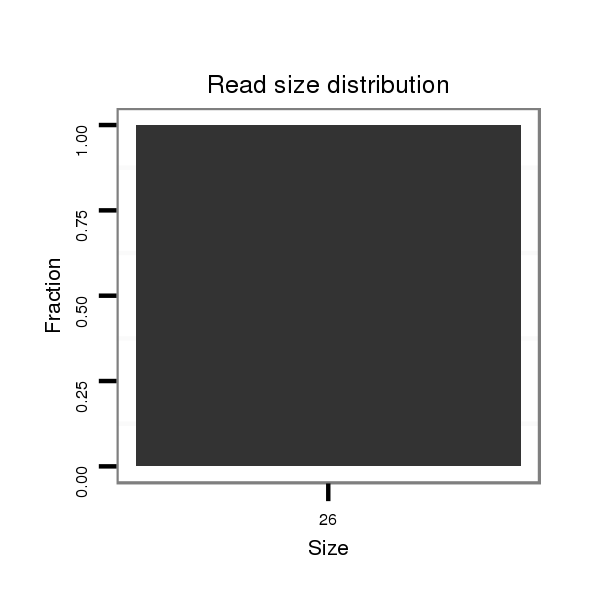
| ...........................................................ACTCAGAACCTGAGACTGTGTCAATC............................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ..............................GTGACAGTGTGTGTCAACCCAGGGC.......................................................................................................................................................... | 25 | 0 | 3 | 0.33 | 1 | 1 |
| .....................TCAGAACCTGTGACAGTGTGTGTCAACC................................................................................................................................................................ | 28 | 0 | 4 | 0.25 | 1 | 1 |
|
CACAGTTGGGTCCCGTCTGTGAGTCTTGGACACTGTCACACACAGTTGGGTCCCGTCTGTGAGTCTTGGACTCTGACACAGTTAGGTCCCATCTGTGAGTCTTGGACTCTGACACAGATAGGTCCCATCTGTGAGTCTTGGACTCCGTCGTATACAGGATTCATGTGTCACCACAGAGTGTTCGGTCACTTTTGTCTATCTTCGGTTTA
***********************************....(((((..((..((((((....(((((...((((....(((((.((((.((((((((((((((...))))....)))))))))).)))).)))))))))...)))))........))))))..)))))))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................GTTCGGTCACTTTTGTCTATCTTCGGTT.. | 28 | 0 | 1 | 2.00 | 2 | 2 |
| ...................................................................................................................................................................................GTTCTGTCACTTTTGTCTATCTTCGGTT.. | 28 | 1 | 1 | 1.00 | 1 | 1 |
| ...........................................AGTTGGGTCCCGTCTGTGAGTCTTGGAC.......................................................................................................................................... | 28 | 0 | 7 | 0.14 | 1 | 1 |
| ...AGTTGGGTCCCGTCTGTGAGTCTTGGAC.................................................................................................................................................................................. | 28 | 0 | 7 | 0.14 | 1 | 1 |
Generated: 04/30/2015 at 05:49 PM