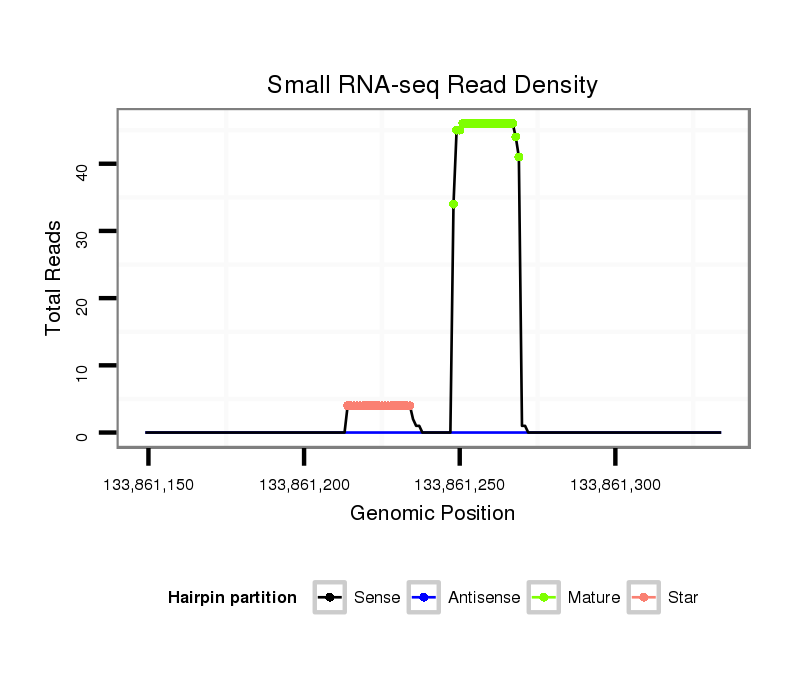
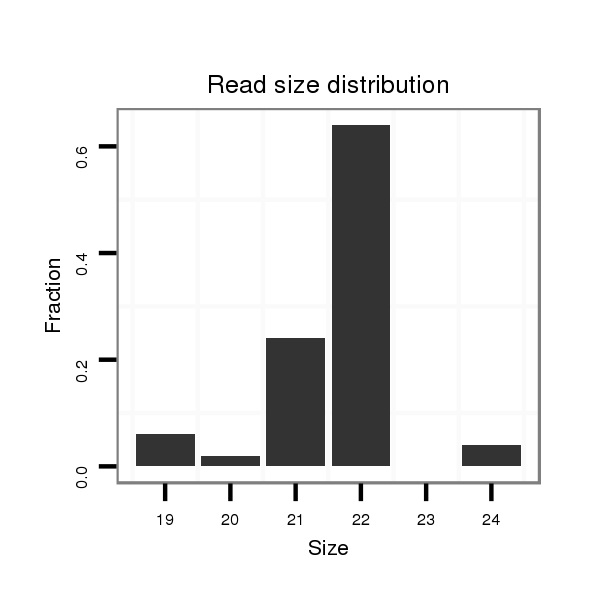
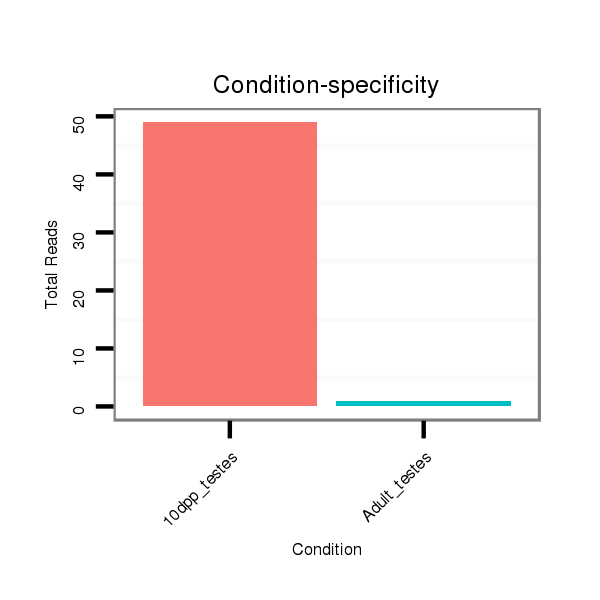
| ...................................................................................................GCACATTACACGGTCGACCTCT................................................................. |
22 |
0 |
1 |
31.00 |
31 |
30 |
1 |
| ....................................................................................................CACATTACACGGTCGACCTCT................................................................. |
21 |
0 |
1 |
8.00 |
8 |
8 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTA................................................................ |
23 |
1 |
1 |
8.00 |
8 |
8 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCA................................................................. |
22 |
1 |
1 |
7.00 |
7 |
7 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTAT............................................................... |
24 |
1 |
1 |
5.00 |
5 |
5 |
0 |
| ....................................................................................................CACATTACACGGTCGACCTTT................................................................. |
21 |
1 |
1 |
2.00 |
2 |
2 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCG.................................................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| ....................................................................................................CACATTACACGGTCGACCTCTAAT.............................................................. |
24 |
3 |
1 |
2.00 |
2 |
2 |
0 |
| ....................................................................................................CACATTACACGGTCGACCTCAAT............................................................... |
23 |
2 |
1 |
2.00 |
2 |
2 |
0 |
| ....................................................................................................CACATTACACGGTCGACCT................................................................... |
19 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTC.................................................................. |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTAG............................................................... |
24 |
2 |
1 |
2.00 |
2 |
2 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCGCAAGA............................................................................................... |
26 |
3 |
1 |
2.00 |
2 |
2 |
0 |
| ....................................................................................................CACATTACACGGTCGACCTC.................................................................. |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCAAT............................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ....................................................................................................CACATTACACGGTAGACCTCT................................................................. |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCACGTTCGCAA................................................................................................. |
24 |
3 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCGCTT................................................................................................. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTAA............................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCGCTAT................................................................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ..................................................................................................AGCACATTACACGGTCGACCTC.................................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCA.................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ....................................................................................................CACATTACACGGTCGACCTCTAA............................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCT.................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ......................................................................................................CATTACACGGTCGACCTCT................................................................. |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTTTAT............................................................. |
26 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTTT............................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| .................................................................AGGTGGTCCGTGGCGCGTTCGC................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTTA................................................................. |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ...................................................................................................GCACATTACACGGTCGACCTCTAC............................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |