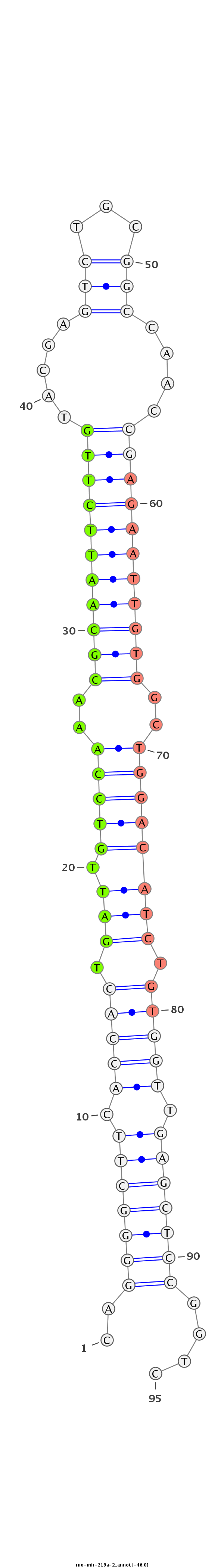
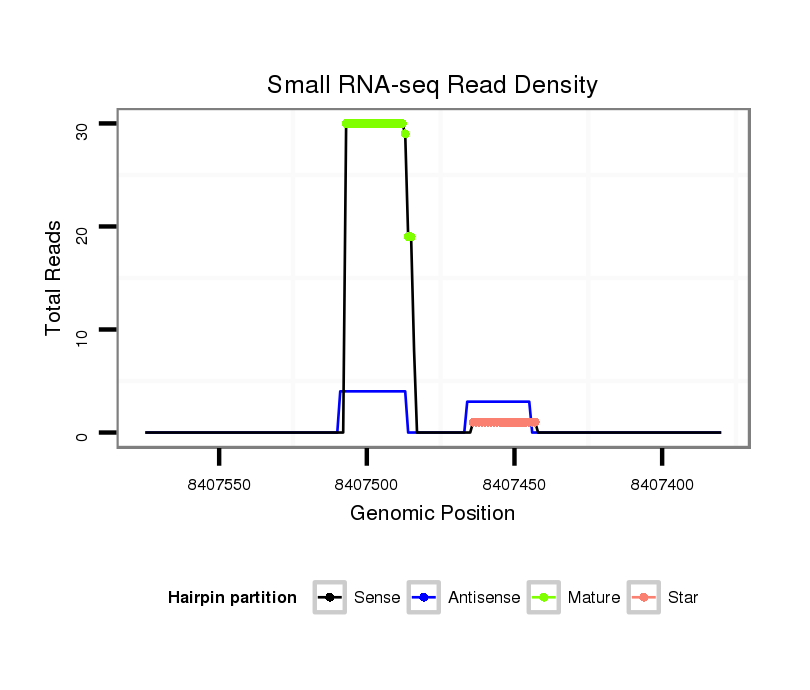

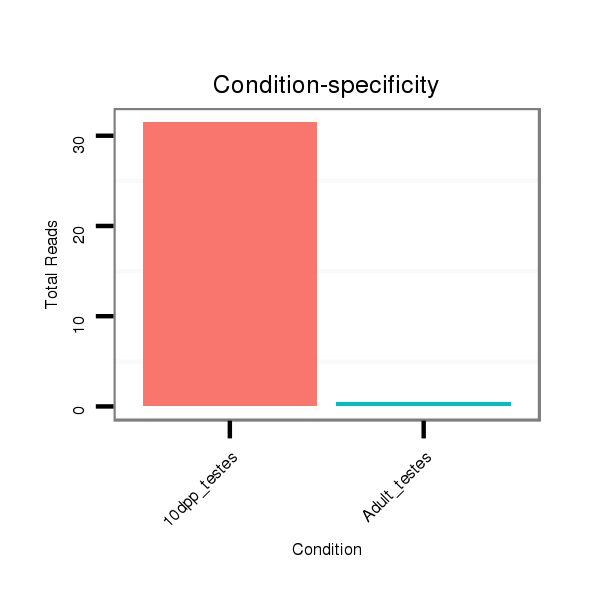
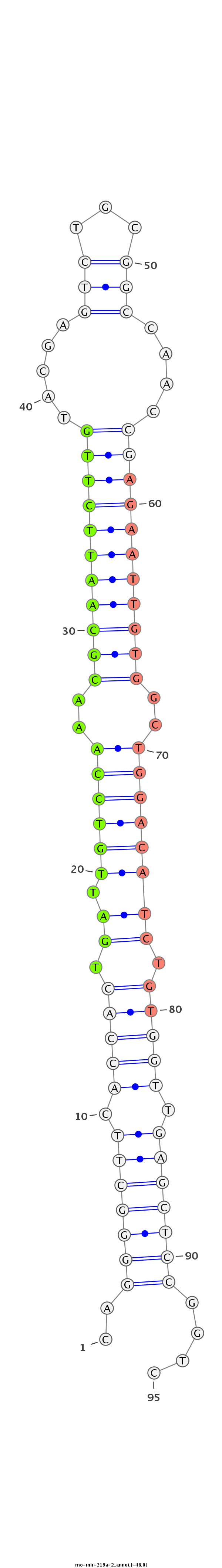
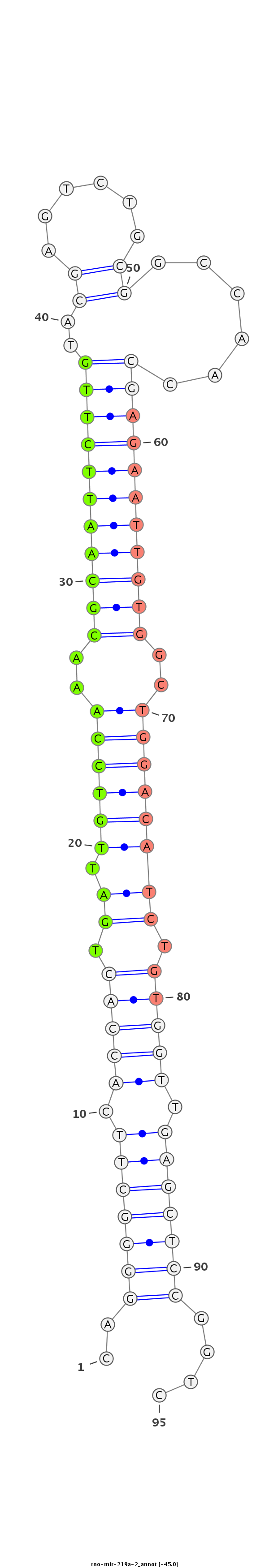
ID:rno-mir-219a-2 |
Coordinate:chr3:8407430-8407525 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -46.0 | -45.0 | -45.0 |
 |
 |
 |
|
GTGCGCTTTTAAGGAGCCAGGCAGAACCCCGAGGGGGACCGGAGCCCTGAACTCAGGGGCTTCACCACTGATTGTCCAAACGCAATTCTTGTACGAGTCTGCGGCCAACCGAGAATTGTGGCTGGACATCTGTGGTTGAGCTCCGGTCTCAACAGAGACGGGGGCCGGGGGCCGCAGAGAGGAGGTCCACTCTGGG
*****************************************************..(((((((.(((((.(((.(((((..(((((((((((.....(((...)))....)))))))))))..)))))))).))))).)))))))....************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
GSM1528817 Adult_testes |
|---|---|---|---|---|---|---|---|
| ....................................................................TGATTGTCCAAACGCAATTCTCGT........................................................................................................ | 24 | 1 | 2 | 12.00 | 24 | 24 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTCTTG......................................................................................................... | 23 | 0 | 1 | 11.00 | 11 | 11 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTCT........................................................................................................... | 21 | 0 | 2 | 10.00 | 20 | 19 | 1 |
| ....................................................................TGATTGTCCAAACGCAATTCTTGT........................................................................................................ | 24 | 0 | 1 | 8.00 | 8 | 8 | 0 |
| ...................................................................ATGATTGTCCAAACGCAATTCTCGT........................................................................................................ | 25 | 2 | 2 | 5.50 | 11 | 1 | 10 |
| ....................................................................TGATTGTCCAAACGCAATTCTAG......................................................................................................... | 23 | 1 | 2 | 2.00 | 4 | 4 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTC............................................................................................................ | 20 | 0 | 2 | 1.00 | 2 | 2 | 0 |
| ...............................................................................................................AGAATTGTGGCTGGACATCTGTAT............................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 |
| ...............................................................................................................AGAATTGTGGCTGGACATCTGT............................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTCTAGT........................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACGCACTTCATG......................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACCCAATTCTTGT........................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTCTTGA........................................................................................................ | 24 | 1 | 2 | 0.50 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACGCAATTCTCGTT....................................................................................................... | 25 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ....................................................................GGATTGTCCAAACGCAATTCTCGT........................................................................................................ | 24 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ....................................................................TGACTGTCCAAACGCAATTCTCGT........................................................................................................ | 24 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAAACGCAATT............................................................................................................. | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| .....................................................................GATTGTCCAAACGCAATTCT........................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ....................................................................TGATTGTCCAACCCCAATTCTAG......................................................................................................... | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 |
|
CACGCGAAAATTCCTCGGTCCGTCTTGGGGCTCCCCCTGGCCTCGGGACTTGAGTCCCCGAAGTGGTGACTAACAGGTTTGCGTTAAGAACATGCTCAGACGCCGGTTGGCTCTTAACACCGACCTGTAGACACCAACTCGAGGCCAGAGTTGTCTCTGCCCCCGGCCCCCGGCGTCTCTCCTCCAGGTGAGACCC
************************************************..(((((((.(((((.(((.(((((..(((((((((((.....(((...)))....)))))))))))..)))))))).))))).)))))))....***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528819 10dpp_testes |
GSM1528818 Adult_testes_Mili_IP |
|---|---|---|---|---|---|---|---|
| ..................................................................TGACTAACAGGTTTGCGTTAAGA........................................................................................................... | 23 | 0 | 1 | 4.00 | 4 | 4 | 0 |
| .............................................................................................................GCTCTTAACACCGACCTGTAGA................................................................. | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 |
| ............................................................................................................AGCTCTTAACACCGACCTGTAGA................................................................. | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 |
| ...............................................................................................................................................GCCAGAGTTGTGTCT...................................... | 15 | 1 | 20 | 0.20 | 4 | 0 | 4 |
Generated: 04/30/2015 at 05:43 PM