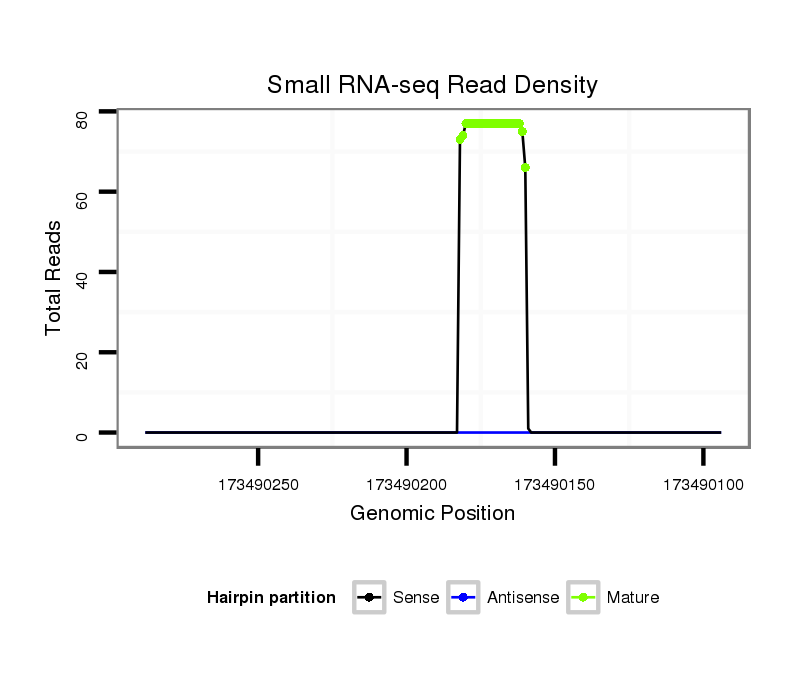
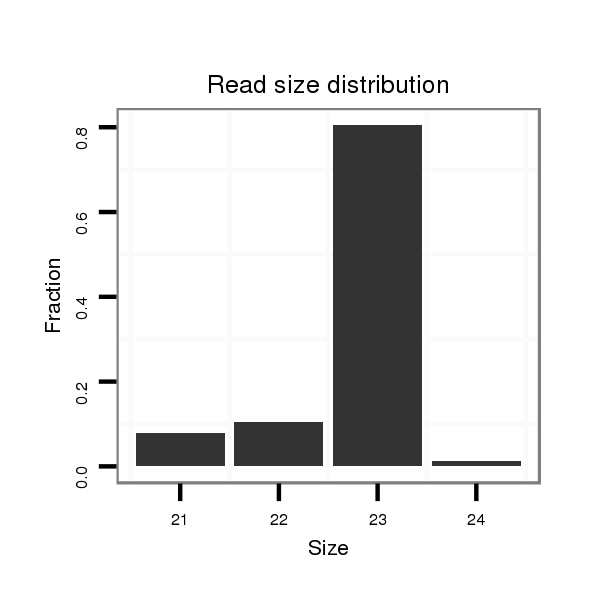
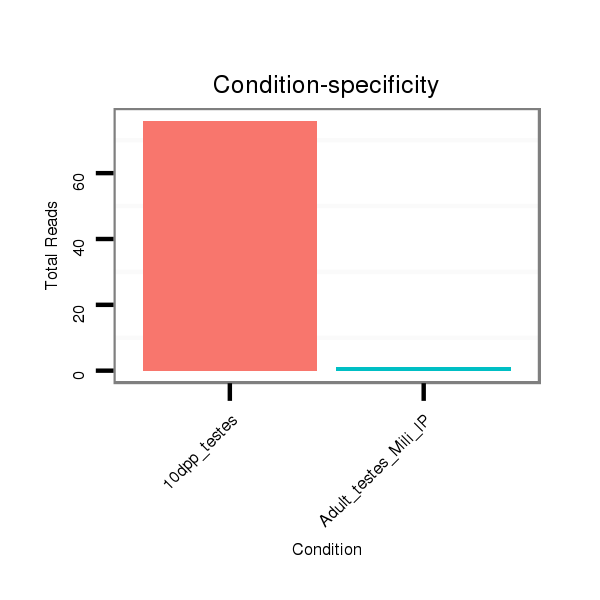
| ..........................................................................................................TAATACTGCCTGGTAATGATGAC.................................................................. |
23 |
0 |
1 |
62.00 |
62 |
62 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGACA................................................................. |
24 |
1 |
1 |
28.00 |
28 |
28 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGACT................................................................. |
24 |
1 |
1 |
16.00 |
16 |
16 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGA................................................................... |
22 |
0 |
1 |
8.00 |
8 |
7 |
1 |
| ...........................................................................................................AATACTGCCTGGTAATGATGACT................................................................. |
23 |
1 |
1 |
4.00 |
4 |
4 |
0 |
| ............................................................................................................ATACTGCCTGGTAATGATGAC.................................................................. |
21 |
0 |
1 |
3.00 |
3 |
3 |
0 |
| ...........................................................................................................AATACTGCCTGGTAATGATGACA................................................................. |
23 |
1 |
1 |
3.00 |
3 |
3 |
0 |
| .........................................................................................................ATAATACTGCCTGGTAATGATGAC.................................................................. |
24 |
1 |
1 |
3.00 |
3 |
3 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATG.................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGAAA................................................................. |
24 |
2 |
1 |
2.00 |
2 |
2 |
0 |
| ..........................................................................................................TAATACTGCCTGGTGATGATGAC.................................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ............................................................................................................ATACTGCCTGGTAATGATAA................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACTGCCTAGTAATGATGACT................................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGACG................................................................. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATTCTGCCTGGTCATGATGAC.................................................................. |
23 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ......................................................................CATCTTACTGGGCAGCATTGGAA...................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACTGCCTGGAAATGATGAC.................................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ...........................................................................................................AATACTGCCTGGTAATGATGA................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGACTTCT.............................................................. |
27 |
3 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACTGCCTGGTAATGATGACTT................................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |
0 |
| ..........................................................................................................TAATACCGCCTGGTAATGATGCC.................................................................. |
23 |
2 |
1 |
1.00 |
1 |
1 |
0 |