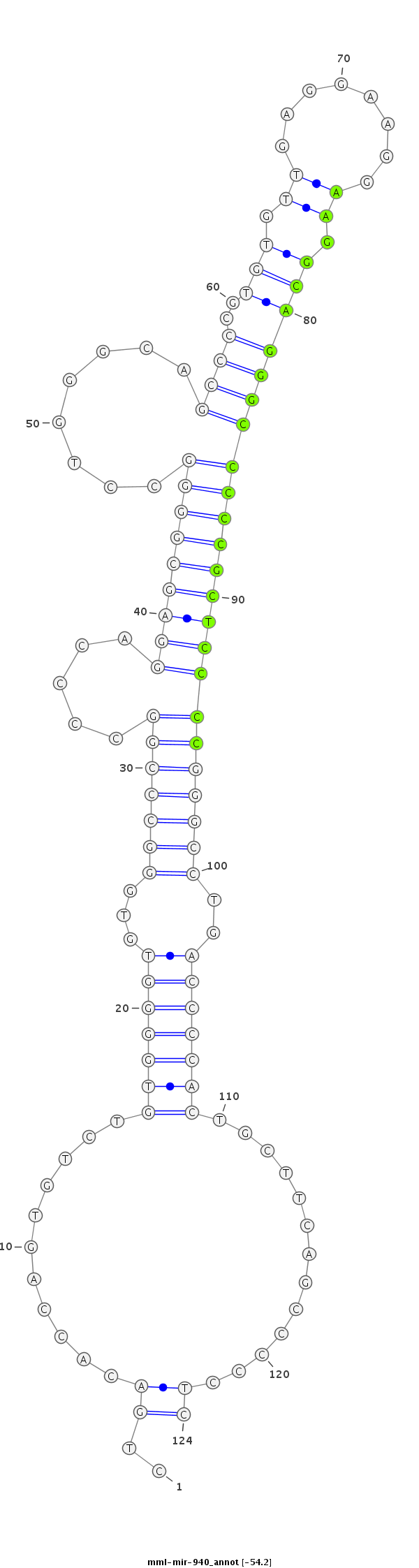
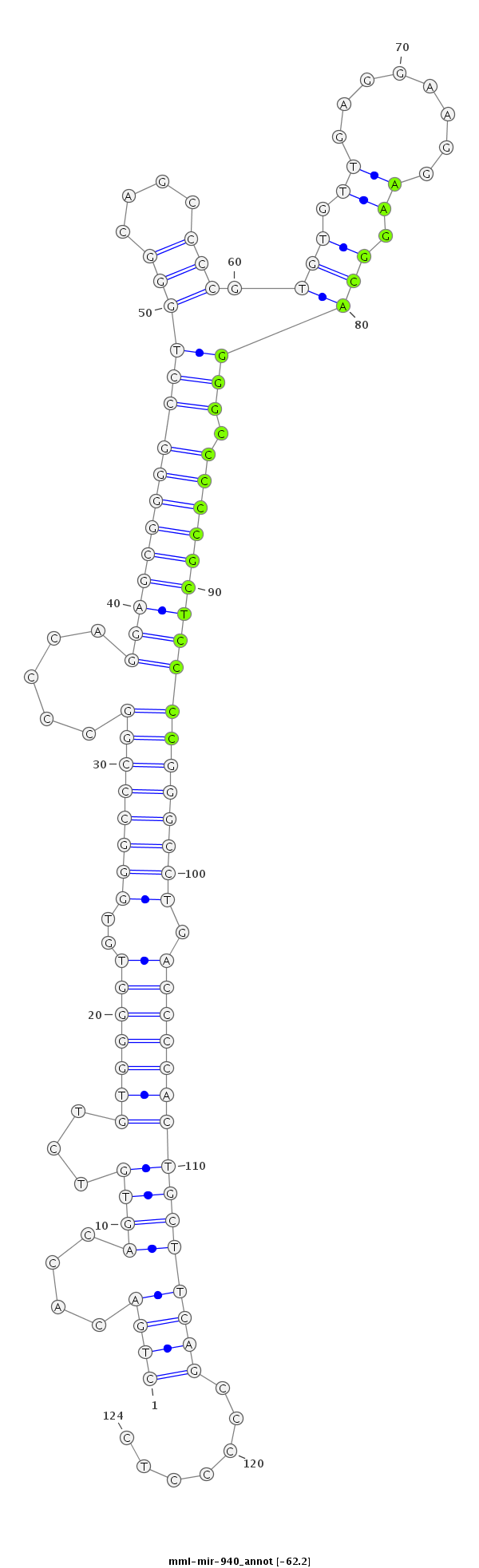
ID:mml-mir-940 |
Coordinate:chr20:2288681-2288774 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -62.2 | -62.2 | -62.2 | -62.2 |
 |
 |
 |
 |
|
GCTGGTCCATGTGGAGCTCCTTCAAGCCGAAGCATCTGACACCAGTGTCTGTGGGGTGTGGGCCCGGCCCCAGGAGCGGGGCCTGGGCAGCCCCGTGTGTTGAGGAAGGAAGGCAGGGCCCCCGCTCCCCGGGCCTGACCCCACTGCTTCAGCCCCCTCCTGCCTGCCTGCAGCCTGGCCTCGAGGGCCCATCA
***********************************..((...........(((((((...(((((((.....(((((((((........((((..(((.((........)).)))))))))))))))))))))))..))))))).............))*********************************** |
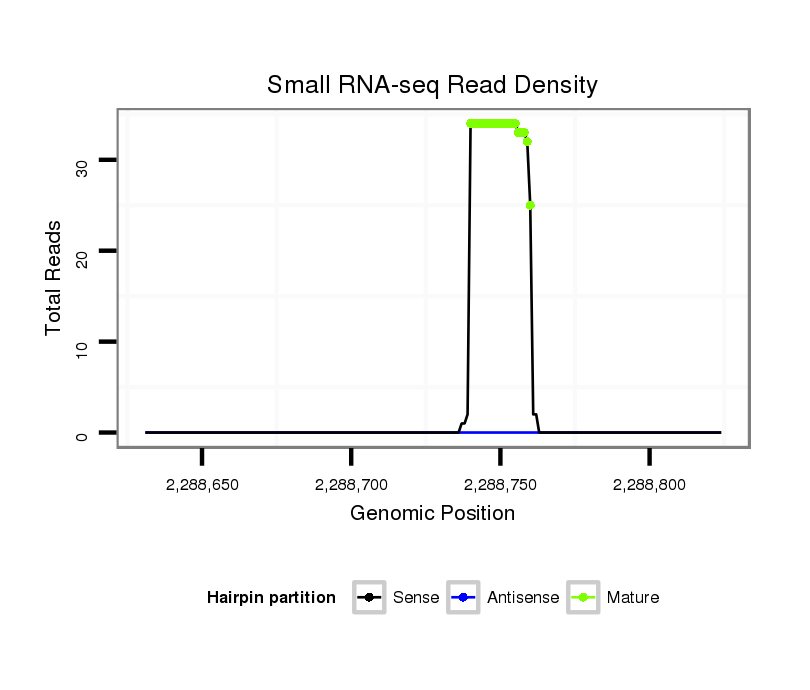
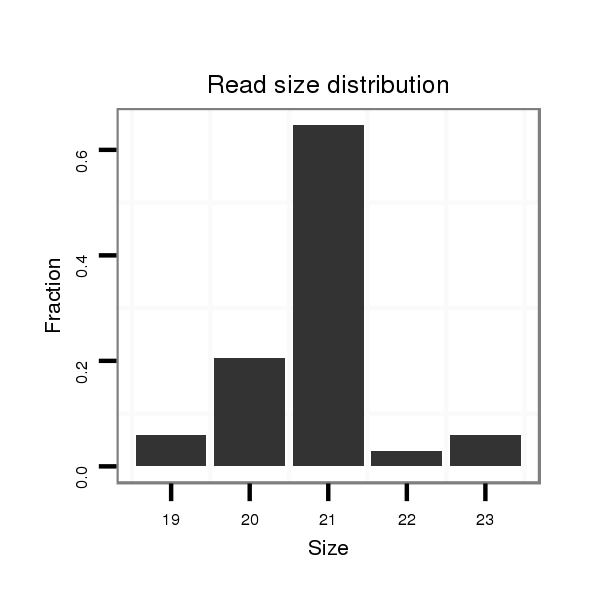
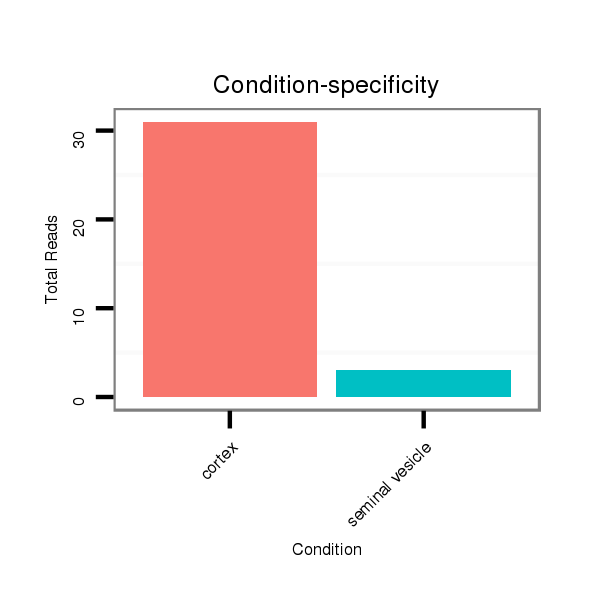
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|---|
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCC................................................................ | 21 | 0 | 1 | 22.00 | 22 | 21 | 1 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCT............................................................... | 22 | 1 | 1 | 11.00 | 11 | 11 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCC................................................................. | 20 | 0 | 1 | 7.00 | 7 | 7 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCTTT............................................................. | 24 | 3 | 1 | 5.00 | 5 | 5 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCTAT............................................................. | 24 | 3 | 2 | 3.00 | 6 | 5 | 1 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCGG.............................................................. | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCTT.............................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCGGTT............................................................ | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................GAAGGCAGGGCCCCCGCTCCCC................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCGGA............................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCA............................................................... | 22 | 1 | 2 | 1.00 | 2 | 2 | 0 |
| ...................................................................................................TTGCGGAAGGAAGGCAGGGCCCCCGC..................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 |
| .............................................................................................................AAGGCAGGGCCGCCGCTCCCC................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................................................................AGGAAGGCAGGGCCCCCGC..................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCC.................................................................. | 19 | 0 | 2 | 1.00 | 2 | 2 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCTAA............................................................. | 24 | 3 | 3 | 0.67 | 2 | 2 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCAT.............................................................. | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCT................................................................ | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCCCA.............................................................. | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCA................................................................. | 20 | 1 | 3 | 0.33 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCAC............................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| .............................................................................................................AAGGCAGGGCCCCCGCTCCCATT.............................................................. | 23 | 3 | 5 | 0.20 | 1 | 1 | 0 |
|
CGACCAGGTACACCTCGAGGAAGTTCGGCTTCGTAGACTGTGGTCACAGACACCCCACACCCGGGCCGGGGTCCTCGCCCCGGACCCGTCGGGGCACACAACTCCTTCCTTCCGTCCCGGGGGCGAGGGGCCCGGACTGGGGTGACGAAGTCGGGGGAGGACGGACGGACGTCGGACCGGAGCTCCCGGGTAGT
***********************************..((...........(((((((...(((((((.....(((((((((........((((..(((.((........)).)))))))))))))))))))))))..))))))).............))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:20 PM