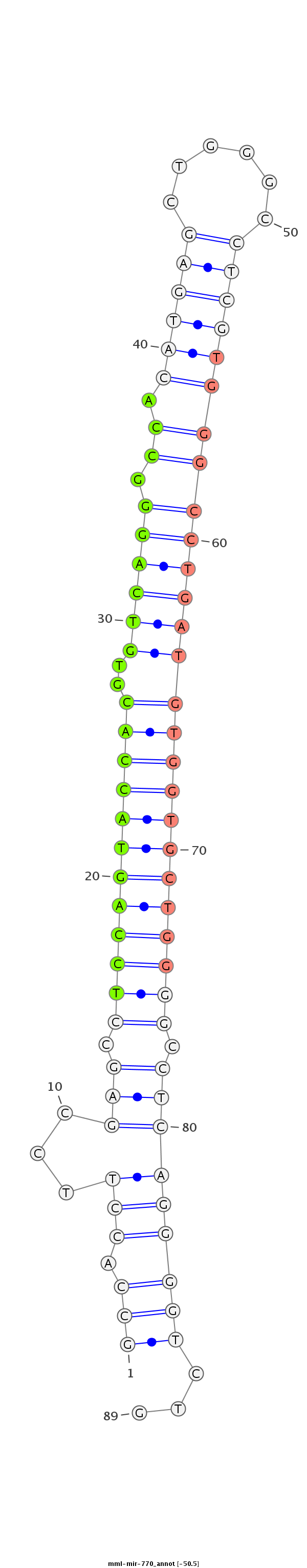
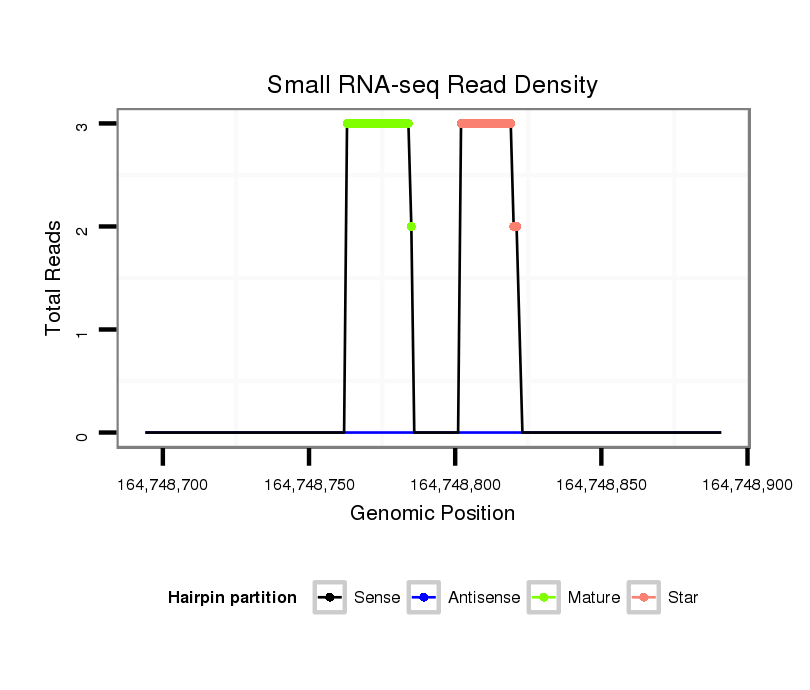
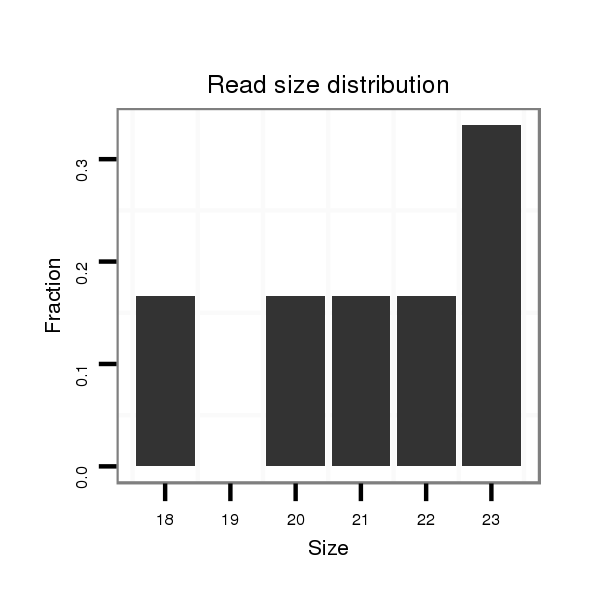
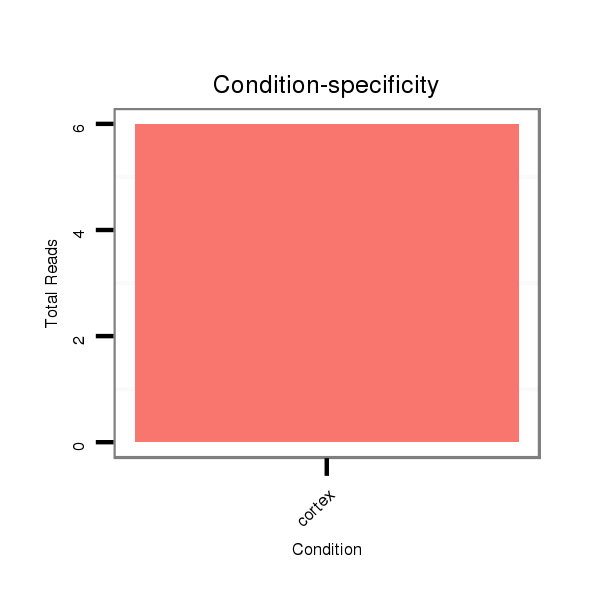
ID:mml-mir-770 |
Coordinate:chr7:164748744-164748841 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
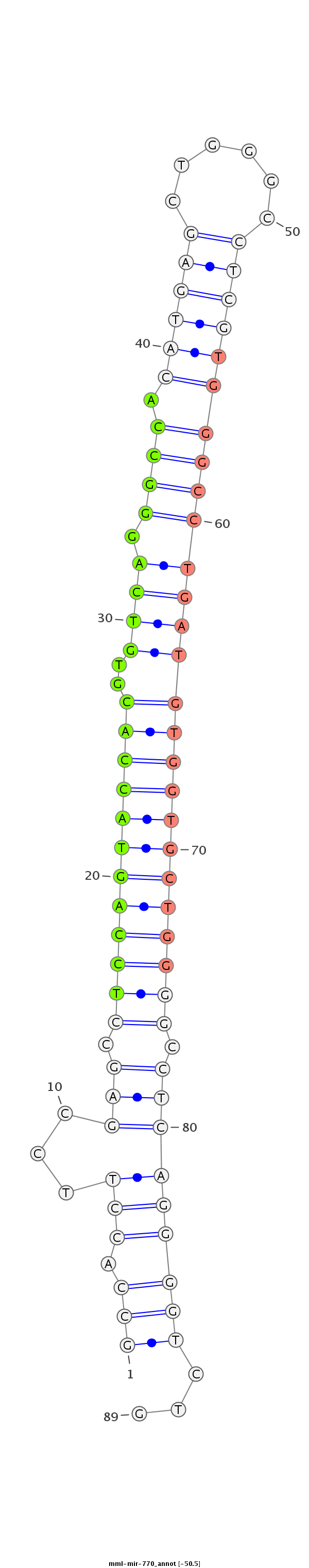
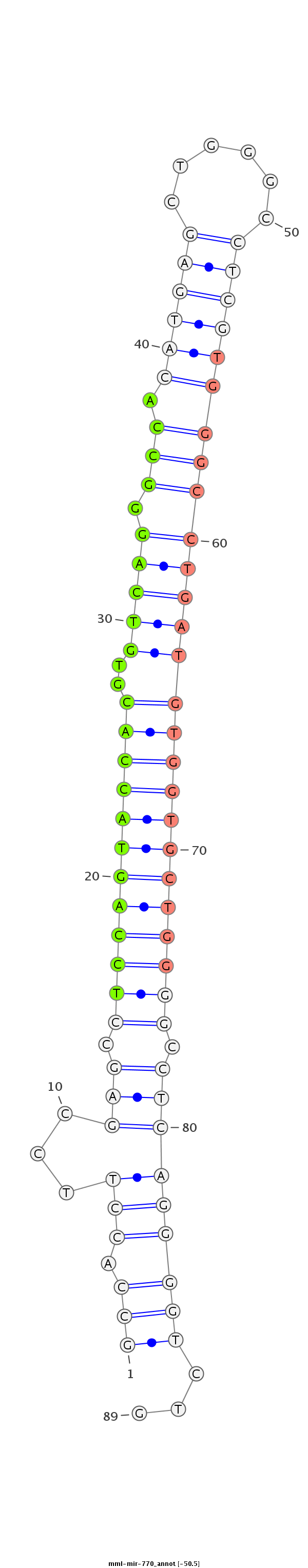
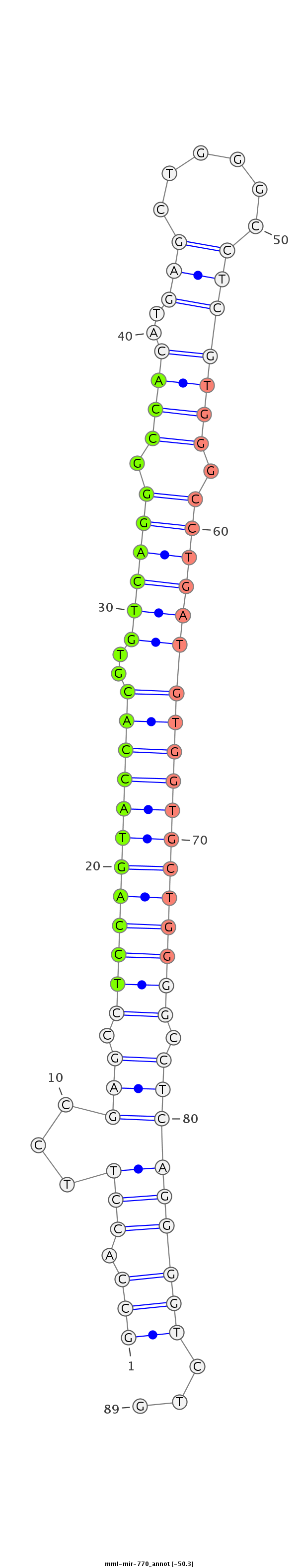
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -50.5 | -50.5 | -50.3 |
 |
 |
 |
|
AGTGGATGGCAACAGCATTTAGAAGGCGGAGGACATGGAATTCATGTGCCAGGAGCCACCTTCCGAGCCTCCAGTACCACGTGTCAGGGCCACATGAGCTGGGCCTCGTGGGCCTGATGTGGTGCTGGGGCCTCAGGGGTCTGCTCTTCTCCTCTTTCAGAATCTGGGGCTGCAGGCTCTGCCTTGGCTGGACTGAGG
******************************************************(((.(((...(((.((((((((((((..((((((.((.((((((......)))))))))))))))))))))))))).)))))))))...******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|
| .............................................................................................................GGGCCTGATGTGGTGCTGGGGCTT................................................................. | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 |
| .....................................................................TCCAGTACCACGTGTCAGGGCCA.......................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 |
| .....................................................................TCCAGTACCACGTGTCAGGGCAG.......................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGG...................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGGT..................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGGGGATC................................................................. | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 |
| .....................................................................TCCAGTACCACGTGTCAGGGCCAA......................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................GGGCCTGATGTGGTGCTGGGGCAT................................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGGG..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................................................TCCAGTACCACGTGTCAGGGCAT.......................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGGGTA................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................................................TCCAGTACCACGTGTCAGGGCC........................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCT........................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................GGGCCTGATGTGGTGCTGGGGTA.................................................................. | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTTGGG.................................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGGAT.................................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| .............................................................................................................GGGCCTGATGTGGTGCTGGGGTAA................................................................. | 24 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ............................................................................................................TGGGCCTGATGTGGTGCTGTTT.................................................................... | 22 | 3 | 20 | 0.15 | 3 | 3 | 0 |
|
TCACCTACCGTTGTCGTAAATCTTCCGCCTCCTGTACCTTAAGTACACGGTCCTCGGTGGAAGGCTCGGAGGTCATGGTGCACAGTCCCGGTGTACTCGACCCGGAGCACCCGGACTACACCACGACCCCGGAGTCCCCAGACGAGAAGAGGAGAAAGTCTTAGACCCCGACGTCCGAGACGGAACCGACCTGACTCC
*******************************************************(((.(((...(((.((((((((((((..((((((.((.((((((......)))))))))))))))))))))))))).)))))))))...****************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:28 PM